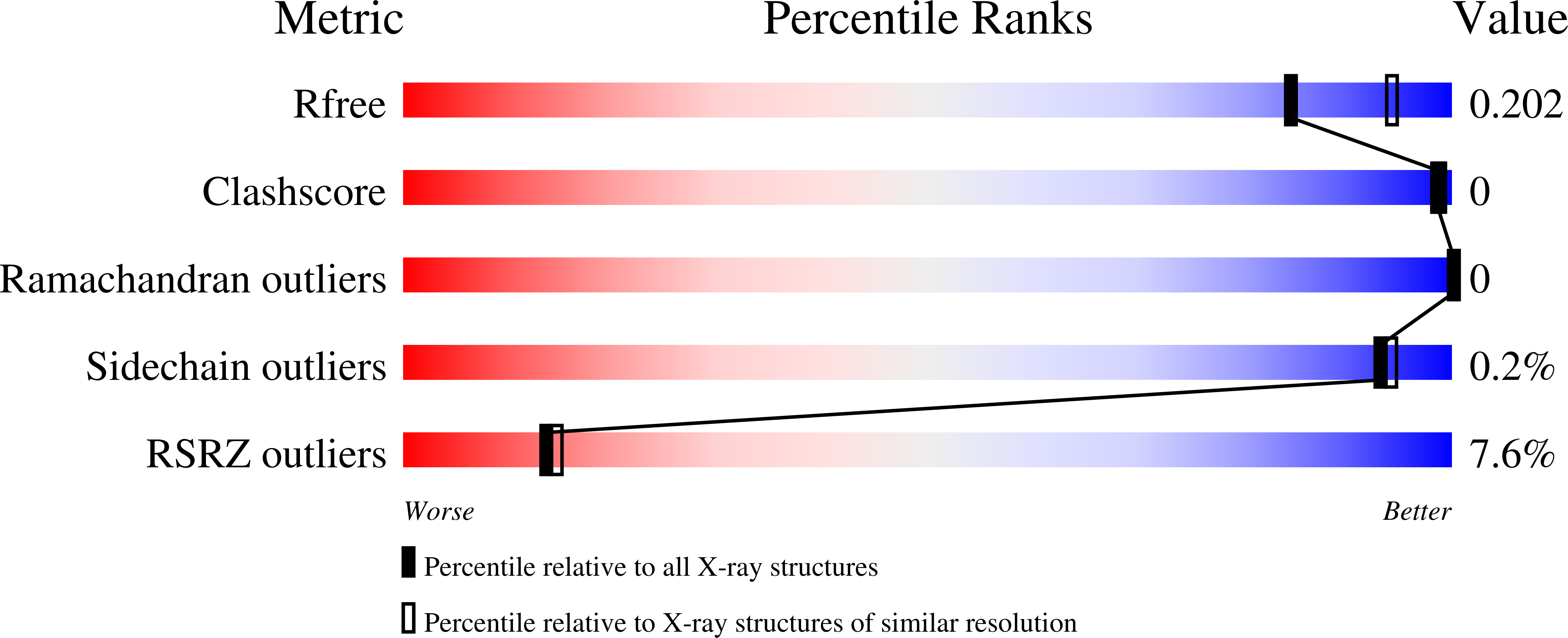
A sugar phosphatase regulates the methylerythritol phosphate (MEP) pathway in malaria parasites.
Guggisberg, A.M., Park, J., Edwards, R.L., Kelly, M.L., Hodge, D.M., Tolia, N.H., Odom, A.R.(2014) Nat Commun 5: 4467-4467
- PubMed: 25058848
- DOI: https://doi.org/10.1038/ncomms5467
- Primary Citation of Related Structures:
4QJB - PubMed Abstract:
Isoprenoid biosynthesis through the methylerythritol phosphate (MEP) pathway generates commercially important products and is a target for antimicrobial drug development. MEP pathway regulation is poorly understood in microorganisms. Here we employ a forward genetics approach to understand MEP pathway regulation in the malaria parasite, Plasmodium falciparum. The antimalarial fosmidomycin inhibits the MEP pathway enzyme deoxyxylulose 5-phosphate reductoisomerase (DXR). Fosmidomycin-resistant P. falciparum are enriched for changes in the PF3D7_1033400 locus (hereafter referred to as PfHAD1), encoding a homologue of haloacid dehalogenase (HAD)-like sugar phosphatases. We describe the structural basis for loss-of-function PfHAD1 alleles and find that PfHAD1 dephosphorylates a variety of sugar phosphates, including glycolytic intermediates. Loss of PfHAD1 is required for fosmidomycin resistance. Parasites lacking PfHAD1 have increased MEP pathway metabolites, particularly the DXR substrate, deoxyxylulose 5-phosphate. PfHAD1 therefore controls substrate availability to the MEP pathway. Because PfHAD1 has homologues in plants and bacteria, other HAD proteins may be MEP pathway regulators.
Organizational Affiliation:
Department of Pediatrics, Washington University School of Medicine, St. Louis, MO 63110, USA.