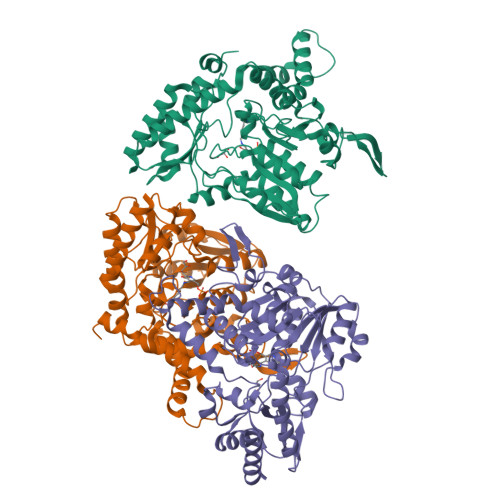
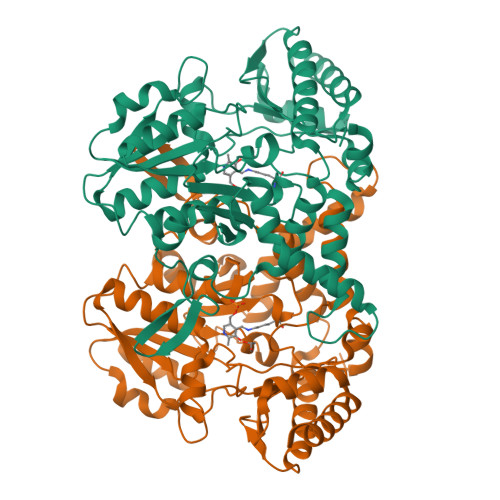
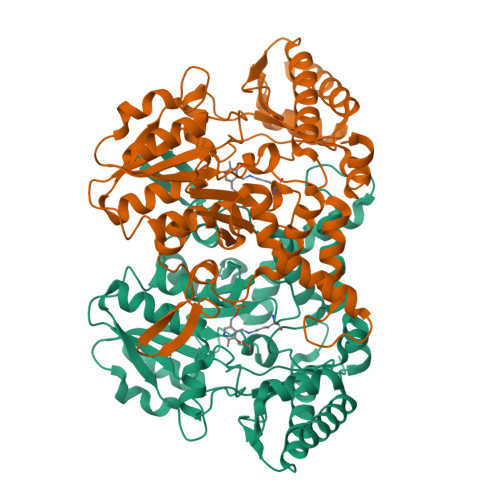
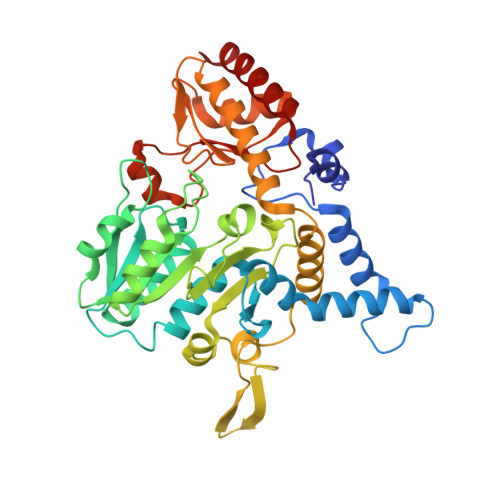
Structural changes during cysteine desulfurase CsdA and sulfur acceptor CsdE interactions provide insight into the trans-persulfuration.
Kim, S., Park, S.(2013) J Biological Chem 288: 27172-27180
- PubMed: 23913692
- DOI: https://doi.org/10.1074/jbc.M113.480277
- Primary Citation of Related Structures:
4LW2, 4LW4 - PubMed Abstract:
In Escherichia coli, three cysteine desulfurases (IscS, SufS, and CsdA) initiate the delivery of sulfur for various biological processes such as the biogenesis of Fe-S clusters. The sulfur generated as persulfide on a cysteine residue of cysteine desulfurases is further transferred to Fe-S scaffolds (e.g. IscU) or to intermediate cysteine-containing sulfur acceptors (e.g. TusA, SufE, and CsdE) prior to its utilization. Here, we report the structures of CsdA and the CsdA-CsdE complex, which provide insight into the sulfur transfer mediated by the trans-persulfuration reaction. Analysis of the structures indicates that the conformational flexibility of the active cysteine loop in CsdE is essential for accepting the persulfide from the cysteine of CsdA. Additionally, CsdA and CsdE invoke a different binding mode than those of previously reported cysteine desulfurase (IscS) and sulfur acceptors (TusA and IscU). Moreover, the conservation of interaction-mediating residues between CsdA/SufS and CsdE/SufE further suggests that the SufS-SufE interface likely resembles that of CsdA and CsdE.
Organizational Affiliation:
School of Systems Biomedical Science, Soongsil University, Seoul 156-743, Korea.