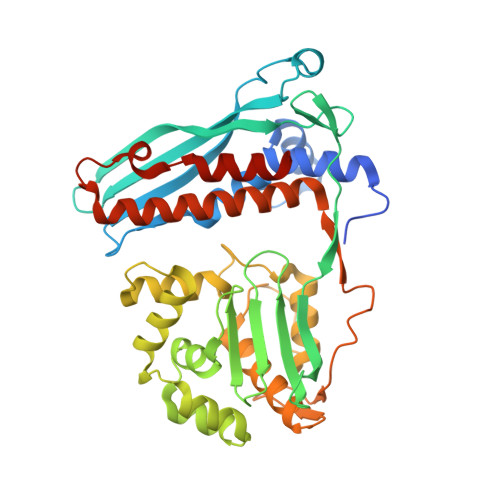
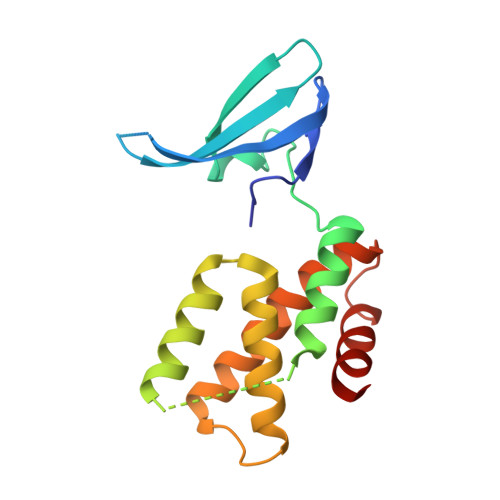
Structure of the Mtb CarD/RNAP beta-Lobes Complex Reveals the Molecular Basis of Interaction and Presents a Distinct DNA-Binding Domain for Mtb CarD.
Gulten, G., Sacchettini, J.C.(2013) Structure 21: 1859-1869
- PubMed: 24055315
- DOI: https://doi.org/10.1016/j.str.2013.08.014
- Primary Citation of Related Structures:
4KBJ, 4KBM - PubMed Abstract:
CarD from Mycobacterium tuberculosis (Mtb) is an essential protein shown to be involved in stringent response through downregulation of rRNA and ribosomal protein genes. CarD interacts with the β-subunit of RNAP and this interaction is vital for Mtb's survival during the persistent infection state. We have determined the crystal structure of CarD in complex with the RNAP β-subunit β1 and β2 domains at 2.1 Å resolution. The structure reveals the molecular basis of CarD/RNAP interaction, providing a basis to further our understanding of RNAP regulation by CarD. The structural fold of the CarD N-terminal domain is conserved in RNAP interacting proteins such as TRCF-RID and CdnL, and displays similar interactions to the predicted homology model based on the TRCF/RNAP β1 structure. Interestingly, the structure of the C-terminal domain, which is required for complete CarD function in vivo, represents a distinct DNA-binding fold.
Organizational Affiliation:
Department of Chemistry, Texas A&M University, College Station, TX 77843, USA.