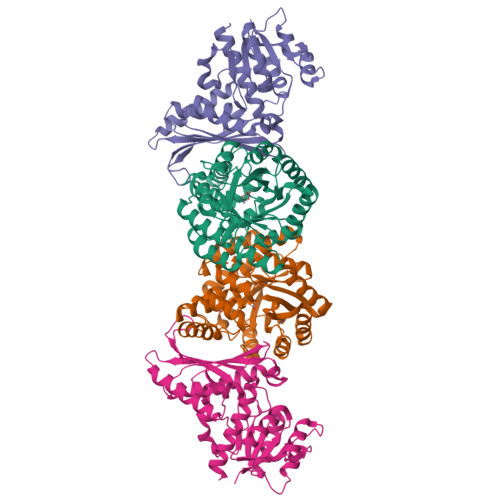
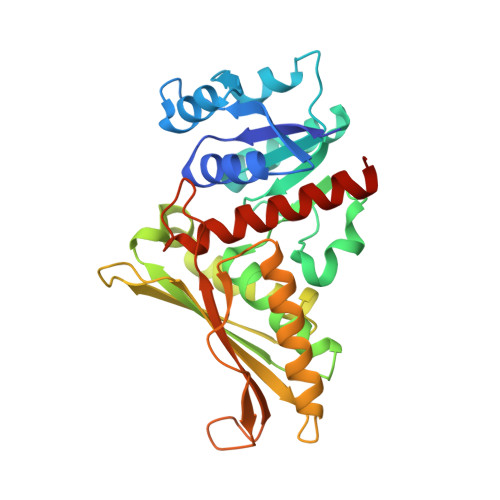
Characterization of an Aldolase-Dehydrogenase Complex from the Cholesterol Degradation Pathway of Mycobacterium tuberculosis.
Carere, J., McKenna, S.E., Kimber, M.S., Seah, S.Y.(2013) Biochemistry 52: 3502-3511
- PubMed: 23614353
- DOI: https://doi.org/10.1021/bi400351h
- Primary Citation of Related Structures:
4JN6 - PubMed Abstract:
HsaF and HsaG are an aldolase and dehydrogenase from the cholesterol degradation pathway of Mycobacterium tuberculosis. HsaF could be heterologously expressed and purified as a soluble dimer, but the enzyme was inactive in the absence of HsaG. HsaF catalyzes the aldol cleavage of 4-hydroxy-2-oxoacids to produce pyruvate and an aldehyde. The enzyme requires divalent metals for activity, with a preference for Mn(2+). The Km values for 4-hydroxy-2-oxoacids were about 20-fold lower than observed for the aldolase homologue, BphI from the polychlorinated biphenyl degradation pathway. Acetaldehyde and propionaldehyde were channeled directly to the dehydrogenase, HsaG, without export to the bulk solvent where they were transformed to acyl-CoA in an NAD(+) and coenzyme A dependent reaction. HsaG is able to utilize aldehydes up to five carbons in length as substrates, with similar catalytic efficiencies. The HsaF-HsaG complex was crystallized and its structure was determined to a resolution of 1.93 Å. Substitution of serine 41 in HsaG with isoleucine or aspartate resulted in about 35-fold increase in Km for CoA but only 4-fold increase in Km dephospho-CoA, suggesting that this residue interacts with the 3'-ribose phosphate of CoA. A second protein annotated as a 4-hydroxy-2-oxopentanoic acid aldolase in M. tuberculosis (MhpE, Rv3469c) was expressed and purified, but was found to lack aldolase activity. Instead this enzyme was found to possess oxaloacetate decarboxylase activity, consistent with the conservation (with the 4-hydroxy-2-oxoacid aldolases) of residues involved in pyruvate enolate stabilization.
Organizational Affiliation:
Department of Molecular and Cellular Biology, University of Guelph , Guelph, Ontario, Canada N1G 2W1.