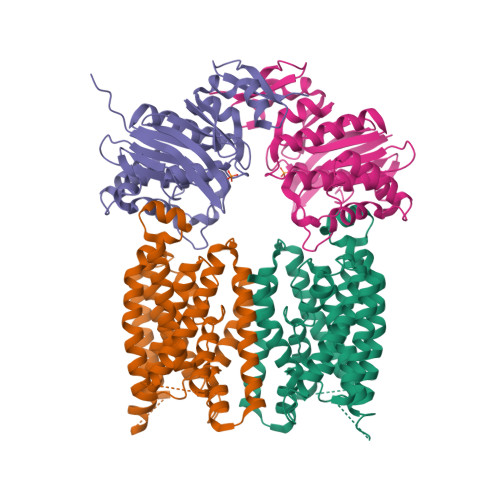
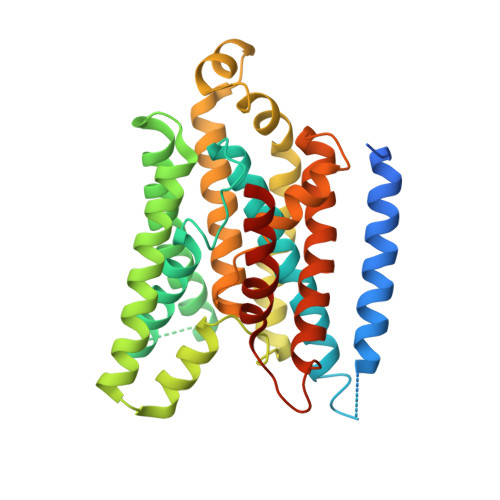
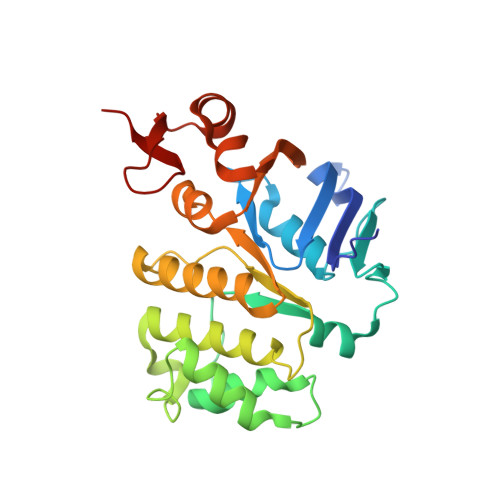
X-ray structure of the Yersinia pestis heme transporter HmuUV.
Woo, J.-S., Zeltina, A., Goetz, B.A., Locher, K.P.(2012) Nat Struct Mol Biol 19: 1310-1315
- PubMed: 23142986
- DOI: https://doi.org/10.1038/nsmb.2417
- Primary Citation of Related Structures:
4G1U - PubMed Abstract:
HmuUV is a bacterial ATP-binding cassette (ABC) transporter that catalyzes heme uptake into the cytoplasm of the gram-negative pathogen Yersinia pestis. We report the crystal structure of HmuUV at 3.0 Å resolution in a nucleotide-free state, which features a heme translocation pathway in an outward-facing conformation, poised to accept a heme from the cognate periplasmic binding protein HmuT. A new assay allowed us to determine in vitro rates of HmuUV-catalyzed heme transport into proteoliposomes and to establish the role of conserved residues in the translocation pathway of HmuUV and at the interface with HmuT. Differences in architecture relative to the related vitamin B(12) transporter BtuCD suggest an adaptation of HmuUV for its smaller substrate. Our study also suggests that type II ABC importers, which include bacterial iron-siderophore, heme and cobalamin transporters, have a coupling mechanism distinct from that of other ABC transporters.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, Eidgenössische Technische Hochschule (ETH) Zürich, CH-8093 Zürich, Switzerland.