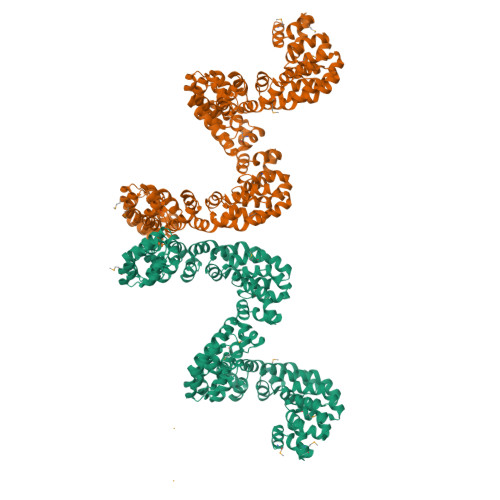
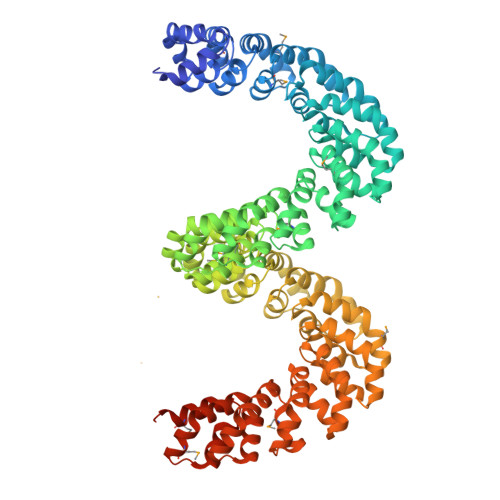
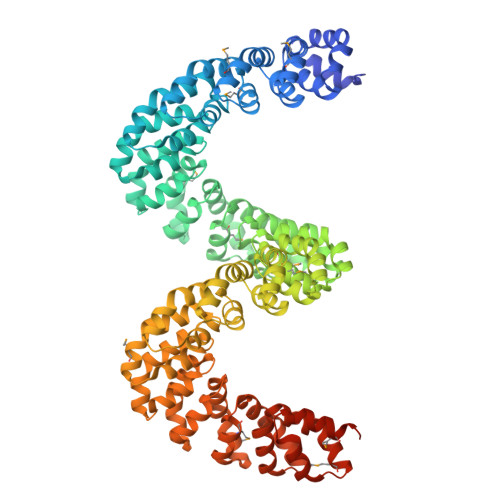
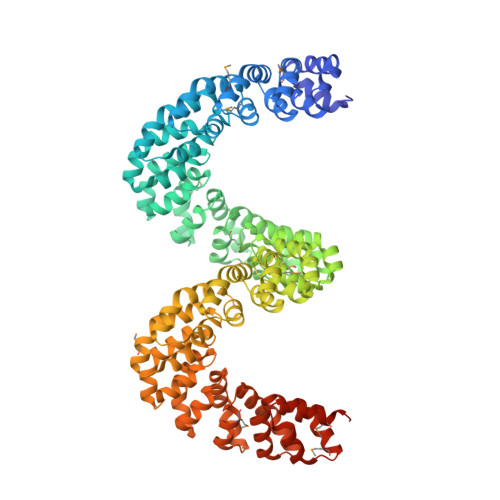Bud, a Helix-Loop-Helix DNA-Binding Domain for Genome Modification
Stella, S., Molina, R., Lopez-Mendez, B., Juillerat, A., Bertonati, C., Daboussi, F., Campos-Olivas, R., Duchateau, P., Montoya, G.(2014) Acta Crystallogr D Biol Crystallogr 70: 2042
- PubMed: 25004980
- DOI: https://doi.org/10.1107/S1399004714011183
- Primary Citation of Related Structures:
4CJ9, 4CJA - PubMed Abstract:
DNA editing offers new possibilities in synthetic biology and biomedicine for modulation or modification of cellular functions to organisms. However, inaccuracy in this process may lead to genome damage. To address this important problem, a strategy allowing specific gene modification has been achieved through the addition, removal or exchange of DNA sequences using customized proteins and the endogenous DNA-repair machinery. Therefore, the engineering of specific protein-DNA interactions in protein scaffolds is key to providing `toolkits' for precise genome modification or regulation of gene expression. In a search for putative DNA-binding domains, BurrH, a protein that recognizes a 19 bp DNA target, was identified. Here, its apo and DNA-bound crystal structures are reported, revealing a central region containing 19 repeats of a helix-loop-helix modular domain (BurrH domain; BuD), which identifies the DNA target by a single residue-to-nucleotide code, thus facilitating its redesign for gene targeting. New DNA-binding specificities have been engineered in this template, showing that BuD-derived nucleases (BuDNs) induce high levels of gene targeting in a locus of the human haemoglobin β (HBB) gene close to mutations responsible for sickle-cell anaemia. Hence, the unique combination of high efficiency and specificity of the BuD arrays can push forward diverse genome-modification approaches for cell or organism redesign, opening new avenues for gene editing.
Organizational Affiliation:
Macromolecular Crystallography Group, Structural Biology and Biocomputing Programme, Spanish National Cancer Research Centre (CNIO), Calle de Melchor Fernández Almagro 3, 28029 Madrid, Spain.