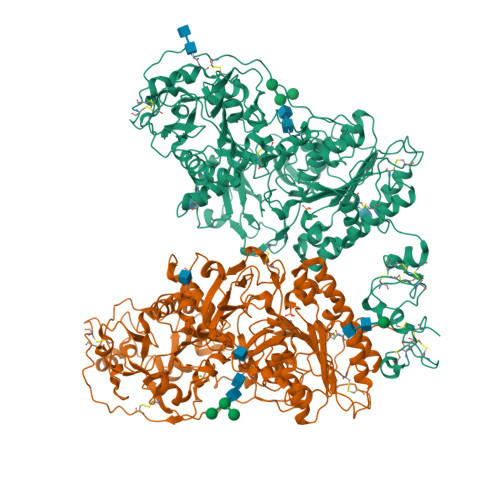
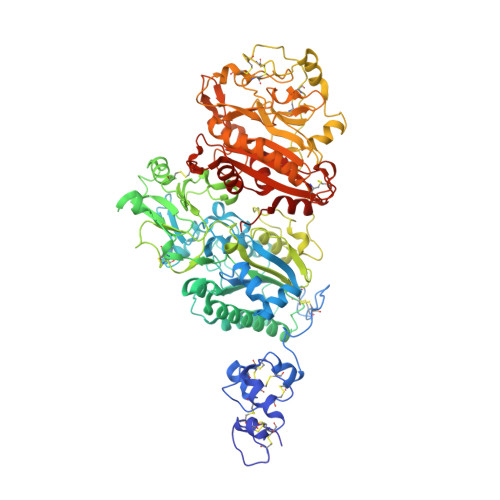
Structure of Npp1, an Ectonucleotide Pyrophosphatase/Phosphodiesterase Involved in Tissue Calcification.
Jansen, S., Perrakis, A., Ulens, C., Winkler, C., Andries, M., Joosten, R.P., Van Acker, M., Luyten, F.P., Moolenaar, W.H., Bollen, M.(2012) Structure 20: 1948
- PubMed: 23041369
- DOI: https://doi.org/10.1016/j.str.2012.09.001
- Primary Citation of Related Structures:
4B56 - PubMed Abstract:
Ectonucleotide pyrophosphatase/phosphodiesterase-1 (NPP1) converts extracellular nucleotides into inorganic pyrophosphate, whereas its close relative NPP2/autotaxin hydrolyzes lysophospholipids. NPP1 regulates calcification in mineralization-competent tissues, and a lack of NPP1 function underlies calcification disorders. Here, we show that NPP1 forms homodimers via intramembrane disulfide bonding, but is also processed intracellularly to a secreted monomer. The structure of secreted NPP1 reveals a characteristic bimetallic active site and a nucleotide-binding groove, but it lacks the lipid-binding pocket and open tunnel present in NPP2. A loop adjacent to the nucleotide-binding site, which is disordered in NPP2, is well ordered in NPP1 and might promote nucleotide binding. Remarkably, the N-terminal somatomedin B-like domains of NPP1, unlike those in NPP2, are flexible and do not contact the catalytic domain. Our results provide a structural basis for the nucleotide pyrophosphatase activity of NPP1 and help to understand how disease-causing mutations may affect NPP1 structure and function.
Organizational Affiliation:
Laboratory of Biosignaling and Therapeutics, Department of Cellular and Molecular Medicine, University of Leuven, 3000 Leuven, Belgium.