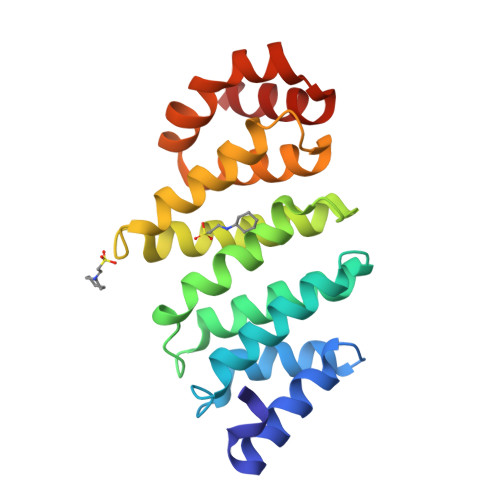
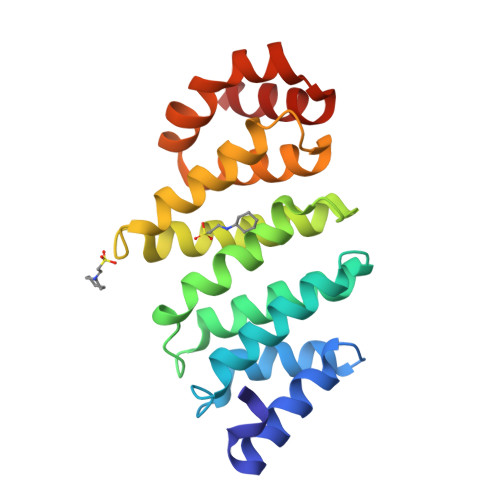
TFIID Taf6-Taf9 Complex Formation Involves the Heat Repeat-Containing C-Terminal Domain of Taf6 and is Modulated by Taf5 Protein.
Scheer, E., Delbac, F., Tora, L., Moras, D., Romier, C.(2012) J Biological Chem 287: 27580
- PubMed: 22696218
- DOI: https://doi.org/10.1074/jbc.M112.379206
- Primary Citation of Related Structures:
4ATG - PubMed Abstract:
The general transcription factor TFIID recognizes specifically the core promoter of genes transcribed by eukaryotic RNA polymerase II, nucleating the assembly of the preinitiation complex at the transcription start site. However, the understanding in molecular terms of TFIID assembly and function remains poorly understood. Histone fold motifs have been shown to be extremely important for the heterodimerization of many TFIID subunits. However, these subunits display several evolutionary conserved noncanonical features when compared with histones, including additional regions whose role is unknown. Here we show that the conserved additional C-terminal region of TFIID subunit TAF6 can be divided into two domains: a small middle domain (TAF6M) and a large C-terminal domain (TAF6C). Our crystal structure of the TAF6C domain from Antonospora locustae at 1.9 Å resolution reveals the presence of five conserved HEAT repeats. Based on these data, we designed several mutants that were introduced into full-length human TAF6. Surprisingly, the mutants affect the interaction between TAF6 and TAF9, suggesting that the formation of the complex between these two TFIID subunits do not only depend on their histone fold motifs. In addition, the same mutants affect even more strongly the interaction between TAF6 and TAF9 in the context of a TAF5-TAF6-TAF9 complex. Expression of these mutants in HeLa cells reveals that most of them are unstable, suggesting their poor incorporation within endogenous TFIID. Taken together, our results suggest that the conserved additional domains in histone fold-containing subunits of TFIID and of co-activator SAGA are important for the assembly of these complexes.
Organizational Affiliation:
Département de Biologie Intégrative, Institut de Génétique et Biologie Moléculaire et Cellulaire (IGBMC), Université de Strasbourg (UDS), CNRS, INSERM, 1 rue Laurent Fries, B.P. 10142, 67404 Illkirch Cedex, France.