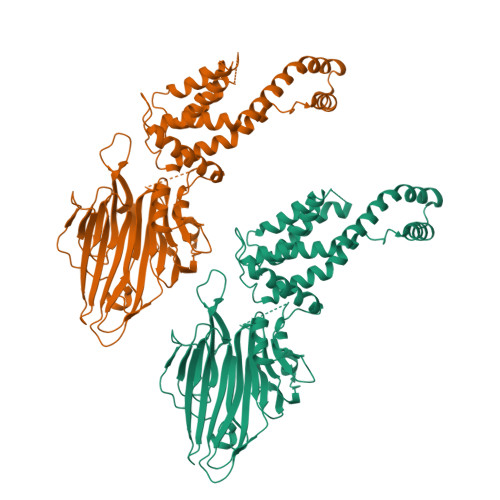
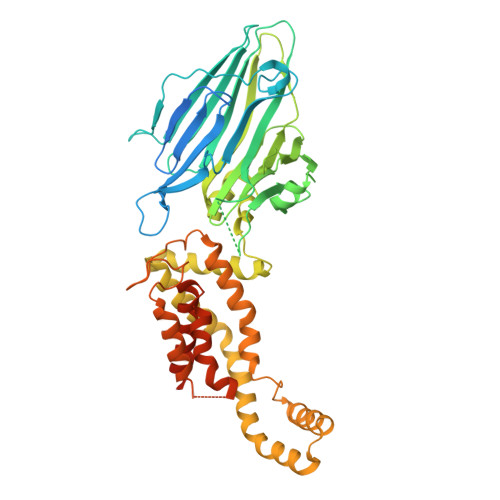
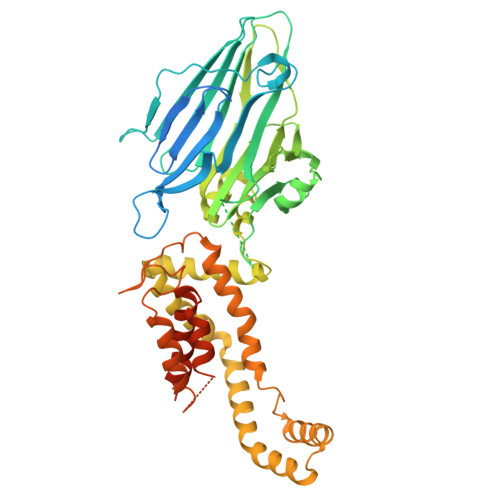
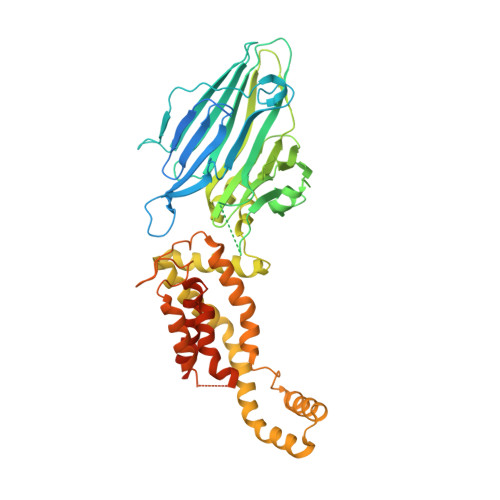
Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase
Kumazaki, K., Kishimoto, T., Furukawa, A., Mori, H., Tanaka, Y., Dohmae, N., Ishitani, R., Tsukazaki, T., Nureki, O.(2014) Sci Rep 4: 7299-7299
- PubMed: 25466392
- DOI: https://doi.org/10.1038/srep07299
- Primary Citation of Related Structures:
3WVF - PubMed Abstract:
Bacterial YidC, an evolutionally conserved membrane protein, functions as a membrane protein chaperone in cooperation with the Sec translocon and as an independent insertase for membrane proteins. In Gram-negative bacteria, the transmembrane and periplasmic regions of YidC interact with the Sec proteins, forming a multi-protein complex for Sec-dependent membrane protein integration. Here, we report the crystal structure of full-length Escherichia coli YidC. The structure reveals that a hydrophilic groove, formed by five transmembrane helices, is a conserved structural feature of YidC, as compared to the previous YidC structure from Bacillus halodurans, which lacks a periplasmic domain. Structural mapping of the substrate- or Sec protein-contact sites suggested the importance of the groove for the YidC functions as a chaperone and an insertase, and provided structural insight into the multi-protein complex.
Organizational Affiliation:
1] Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan [2] Global Research Cluster, RIKEN, 2-1 Hirosawa, Wako-shi, Saitama 351-0198, Japan.