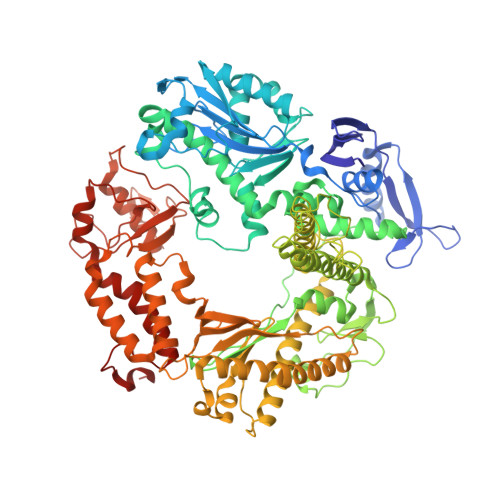
A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex.
Aller, P., Duclos, S., Wallace, S.S., Doublie, S.(2011) J Mol Biol 412: 22-34
- PubMed: 21781974
- DOI: https://doi.org/10.1016/j.jmb.2011.07.007
- PubMed Abstract:
Thymine glycol (Tg) is the most common oxidation product of thymine and is known to be a strong block to replicative DNA polymerases. A previously solved structure of the bacteriophage RB69 DNA polymerase (RB69 gp43) in complex with Tg in the sequence context 5'-G-Tg-G shed light on how Tg blocks primer elongation: The protruding methyl group of the oxidized thymine displaces the adjacent 5'-G, which can no longer serve as a template for primer elongation [Aller, P., Rould, M. A., Hogg, M, Wallace, S. S. & Doublié S. (2007). A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. USA, 104, 814-818.]. Several studies showed that in the sequence context 5'-C-Tg-purine, Tg is more likely to be bypassed by Klenow fragment, an A-family DNA polymerase. We set out to investigate the role of sequence context in Tg bypass in a B-family polymerase and to solve the crystal structures of the bacteriophage RB69 DNA polymerase in complex with Tg-containing DNA in the three remaining sequence contexts: 5'-A-Tg-G, 5'-T-Tg-G, and 5'-C-Tg-G. A combination of several factors-including the associated exonuclease activity, the nature of the 3' and 5' bases surrounding Tg, and the cis-trans interconversion of Tg-influences Tg bypass. We also visualized for the first time the structure of a well-ordered exonuclease complex, allowing us to identify and confirm the role of key residues (Phe123, Met256, and Tyr257) in strand separation and in the stabilization of the primer strand in the exonuclease site.
Organizational Affiliation:
Department of Microbiology andMolecular Genetics, Stafford Hall, University of Vermont, Burlington, VT 05405, USA.