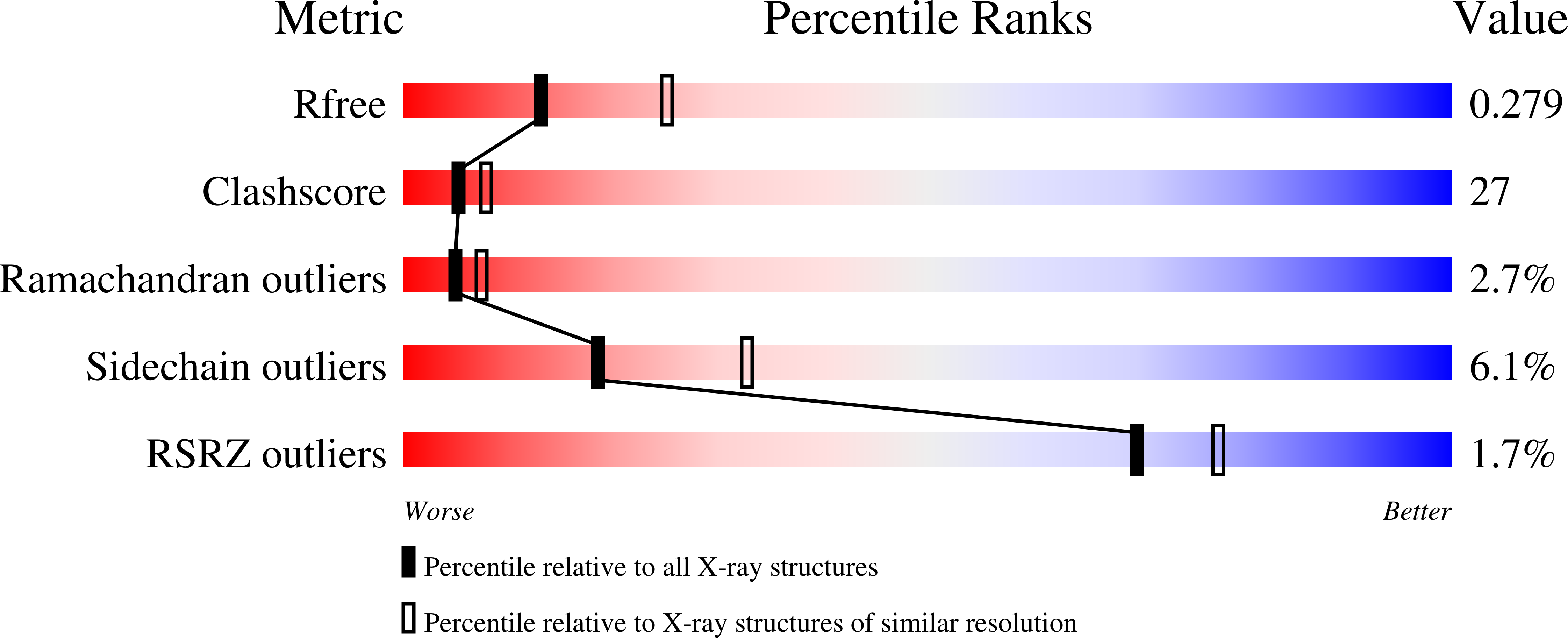
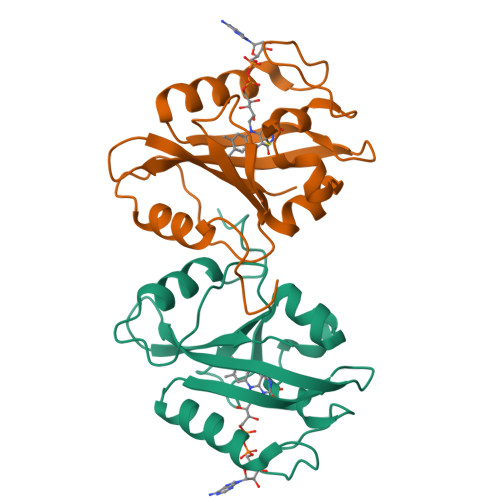
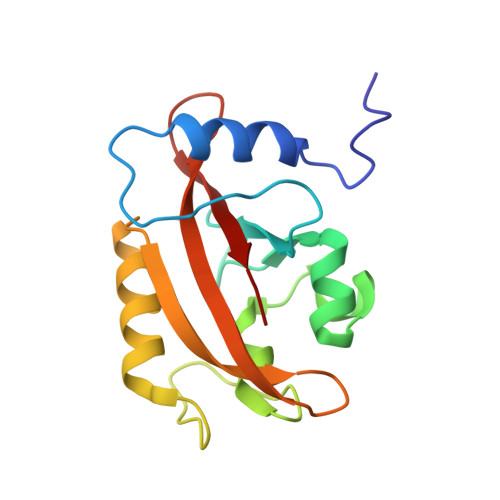
Structure of a Light-Activated LOV Protein Dimer That Regulates Transcription.
Vaidya, A.T., Chen, C.H., Dunlap, J.C., Loros, J.J., Crane, B.R.(2011) Sci Signal 4: ra50-ra50
- PubMed: 21868352
- DOI: https://doi.org/10.1126/scisignal.2001945
- Primary Citation of Related Structures:
3RH8 - PubMed Abstract:
Light, oxygen, or voltage (LOV) protein domains are present in many signaling proteins in bacteria, archaea, protists, plants, and fungi. The LOV protein VIVID (VVD) of the filamentous fungus Neurospora crassa enables the organism to adapt to constant or increasing amounts of light and facilitates proper entrainment of circadian rhythms. Here, we determined the crystal structure of the fully light-adapted VVD dimer and reveal the mechanism by which light-driven conformational change alters the oligomeric state of the protein. Light-induced formation of a cysteinyl-flavin adduct generated a new hydrogen bond network that released the amino (N) terminus from the protein core and restructured an acceptor pocket for binding of the N terminus on the opposite subunit of the dimer. Substitution of residues critical for the switch between the monomeric and the dimeric states of the protein had profound effects on light adaptation in Neurospora. The mechanism of dimerization of VVD provides molecular details that explain how members of a large family of photoreceptors convert light responses to alterations in protein-protein interactions.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA.