Structural and Functional Analyses Reveal That Staphylococcus aureus Antibiotic Resistance Factor HmrA Is a Zinc-dependent Endopeptidase.
Botelho, T.O., Guevara, T., Marrero, A., Arede, P., Fluxa, V.S., Reymond, J.L., Oliveira, D.C., Gomis-Ruth, F.X.(2011) J Biological Chem 286: 25697-25709
- PubMed: 21622555
- DOI: https://doi.org/10.1074/jbc.M111.247437
- Primary Citation of Related Structures:
3RAM - PubMed Abstract:
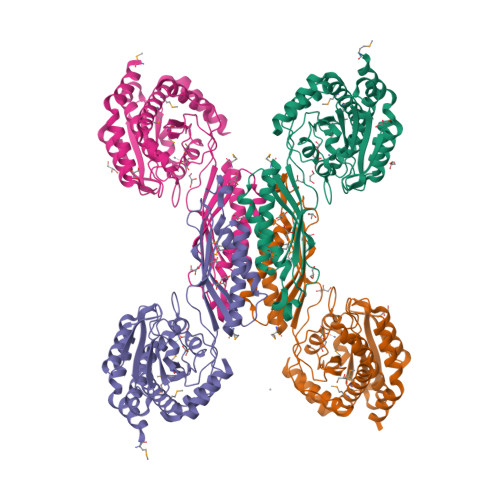
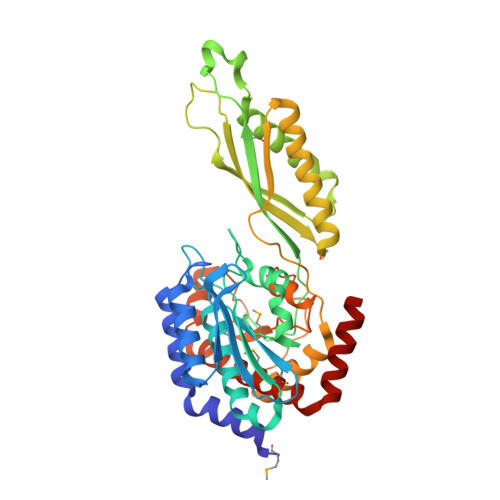
HmrA is an antibiotic resistance factor of methicillin-resistant Staphylococcus aureus. Molecular analysis of this protein revealed that it is not a muramidase or β-lactamase but a nonspecific double-zinc endopeptidase consisting of a catalytic domain and an inserted oligomerization domain, which probably undergo a relative interdomain hinge rotation upon substrate binding. The active-site cleft is located at the domain interface. Four HmrA protomers assemble to a large ∼170-kDa homotetrameric complex of 125 Å. All four active sites are fully accessible and ∼50-70 Å apart, far enough apart to act on a large meshwork substrate independently but simultaneously. In vivo studies with four S. aureus strains of variable resistance levels revealed that the extracellular addition of HmrA protects against loss of viability in the presence of oxacillin and that this protection depends on proteolytic activity. All of these results indicate that HmrA is a peptidase that participates in resistance mechanisms in vivo in the presence of β-lactams. Furthermore, our results have implications for most S. aureus strains of known genomic sequences and several other cocci and bacilli, which harbor close orthologs. This suggests that HmrA may be a new widespread antibiotic resistance factor in bacteria.
Organizational Affiliation:
Proteolysis Laboratory, Department of Structural Biology, Molecular Biology Institute of Barcelona, Barcelona Science Park, Barcelona, Spain.