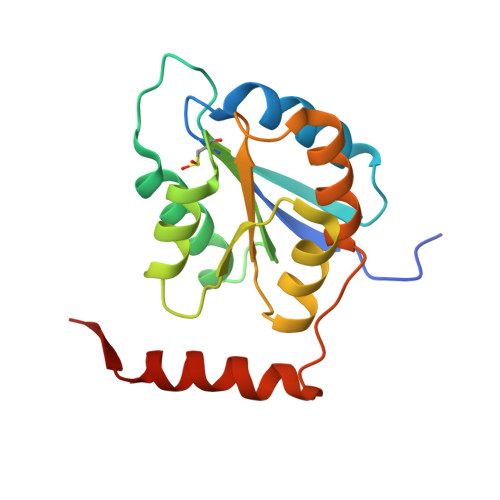
Structural characterization of a ribose-5-phosphate isomerase B from the pathogenic fungus Coccidioides immitis.
Edwards, T.E., Abramov, A.B., Smith, E.R., Baydo, R.O., Leonard, J.T., Leibly, D.J., Thompkins, K.B., Clifton, M.C., Gardberg, A.S., Staker, B.L., Van Voorhis, W.C., Myler, P.J., Stewart, L.J.(2011) BMC Struct Biol 11: 39-39
- PubMed: 21995815
- DOI: https://doi.org/10.1186/1472-6807-11-39
- Primary Citation of Related Structures:
3QD5, 3SDW, 3SGW - PubMed Abstract:
Ribose-5-phosphate isomerase is an enzyme that catalyzes the interconversion of ribose-5-phosphate and ribulose-5-phosphate. This family of enzymes naturally occurs in two distinct classes, RpiA and RpiB, which play an important role in the pentose phosphate pathway and nucleotide and co-factor biogenesis.
Organizational Affiliation:
Seattle Structural Genomics Center for Infectious Disease (SSGCID), USA. tedwards@embios.com