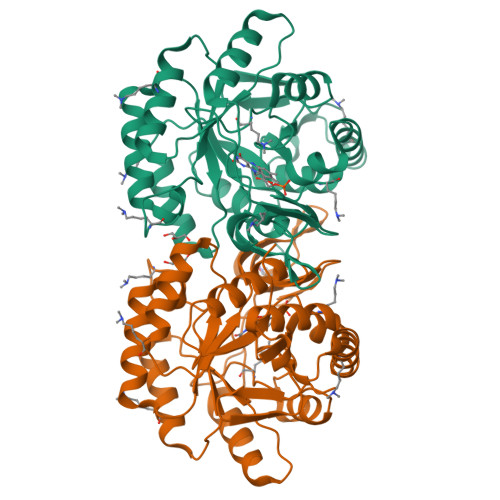
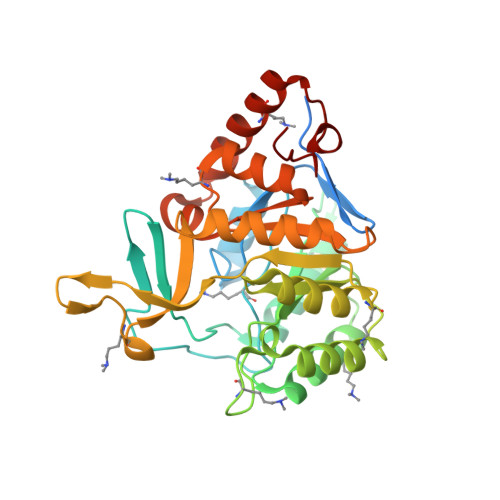
Structure of the putative dihydroorotate dehydrogenase from Streptococcus mutans
Liu, Y., Gao, Z.-Q., Liu, C.-P., Xu, J.-H., Li, L.-F., Ji, C.-N., Su, X.-D., Dong, Y.-H.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 182-187
- PubMed: 21301083
- DOI: https://doi.org/10.1107/S1744309110048414
- Primary Citation of Related Structures:
3OIX - PubMed Abstract:
Streptococcus mutans is one of the pathogenic species involved in dental caries, especially in the initiation and development stages. Here, the crystal structure of SMU.595, a putative dihydroorotate dehydrogenase (DHOD) from S. mutans, is reported at 2.4 Å resolution. DHOD is a flavin mononucleotide-containing enzyme which catalyzes the oxidation of L-dihydroorotate to orotate, which is the fourth step and the only redox reaction in the de novo biosynthesis of pyrimidine nucleotides. The reductive lysine-methylation procedure was applied in order to improve the diffraction qualities of the crystals. Analysis of the S. mutans DHOD crystal structure shows that this enzyme is a class 1A DHOD and also suggests potential sites that could be exploited for the design of highly specific inhibitors using the structure-based chemotherapeutic design technique.
Organizational Affiliation:
Department of Genetics, School of Life Science, Fudan University, Shanghai, People's Republic of China.