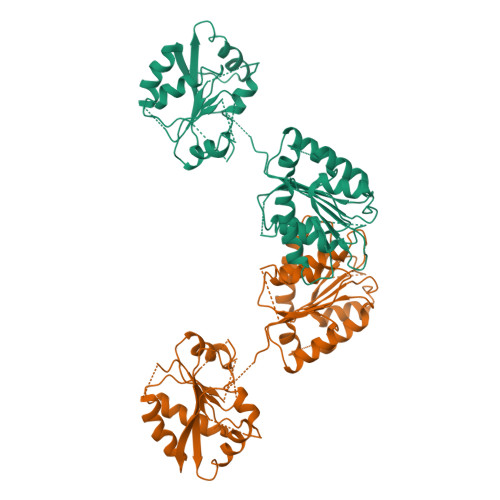
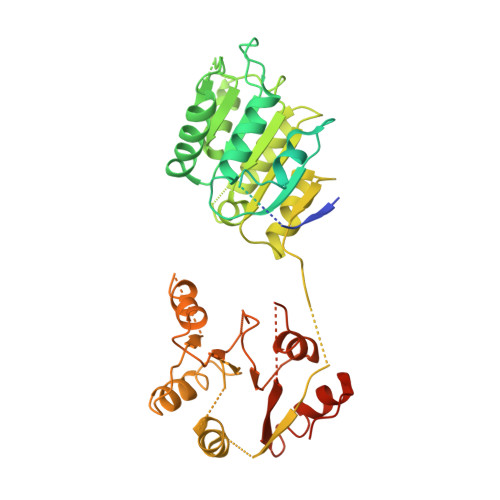
Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
Fan, J.S., Cheng, Z., Zhang, J., Noble, C., Zhou, Z., Song, H., Yang, D.(2009) J Mol Biology 388: 1-10
- PubMed: 19281819
- DOI: https://doi.org/10.1016/j.jmb.2009.03.004
- Primary Citation of Related Structures:
2KBE, 2KBF, 3FHO - PubMed Abstract:
DEAD-box protein 5 (Dbp5p) plays very important roles in RNA metabolism from transcription, to translation, to RNA decay. It is an RNA helicase and functions as an essential RNA export factor from nucleus. Here, we report the solution NMR structures of the N- and C-terminal domains (NTD and CTD, respectively) of Dbp5p from Saccharomyces cerevisiae (ScDbp5p) and X-ray crystal structure of Dbp5p from Schizosaccharomyces pombe (SpDbp5p) in the absence of nucleotides and RNA. The crystal structure clearly shows that SpDbp5p comprises two RecA-like domains that do not interact with each other. NMR results show that the N-terminal flanking region of ScDpbp5 (M1-E70) is intrinsically unstructured and the region Y71-R121 including the Q motif is highly dynamic on millisecond-microsecond timescales in solution. The C-terminal flanking region of ScDbp5p forms a short beta-strand and a long helix. This helix is unique for ScDbp5p and has not been observed in other DEAD-box proteins. Compared with other DEAD-box proteins, Dbp5p has an extra insert with six residues in the CTD. NMR structure reveals that the insert is located in a solvent-exposed loop capable of interacting with other proteins. ATP and ADP titration experiments show that both ADP and ATP bind to the consensus binding site in the NTD of ScDbp5p but do not interact with the CTD at all. Binding of ATP or ADP to NTD induces significant conformational rearrangement too.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543, Singapore.