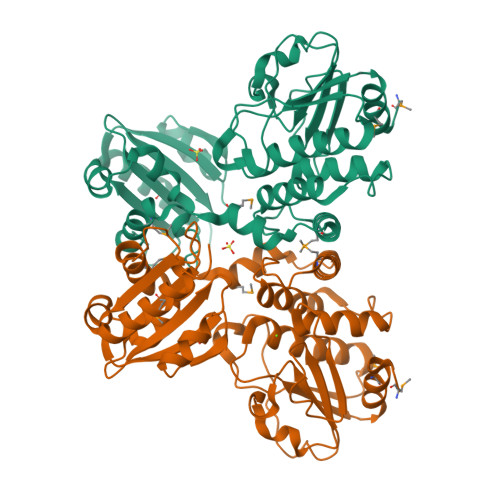
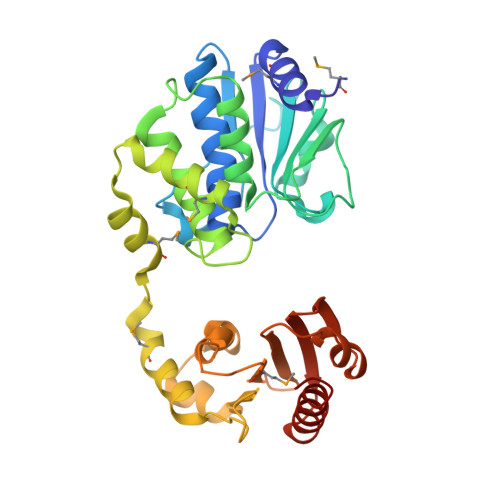
Crystal structure of SH1221 protein from Staphylococcus haemolyticus, Northeast Structural Genomics Consortium Target ShR87
Forouhar, F., Neely, H., Seetharaman, J., Mao, L., Xiao, R., Ciccosanti, C., Foote, E.L., Wang, D., Everett, J.K., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.