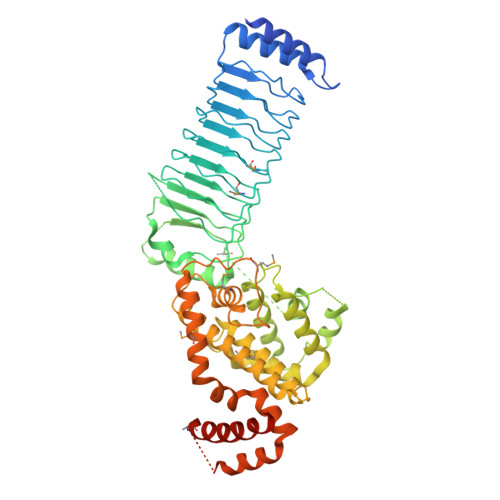
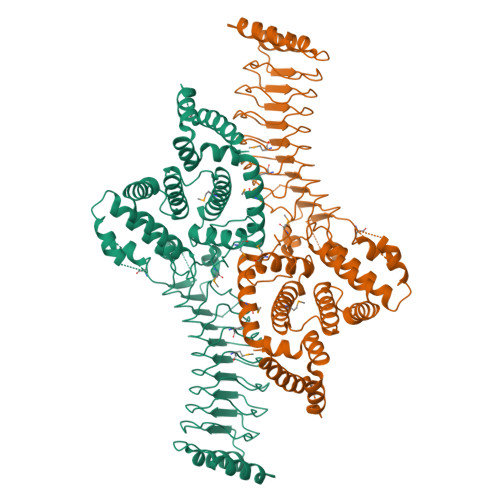
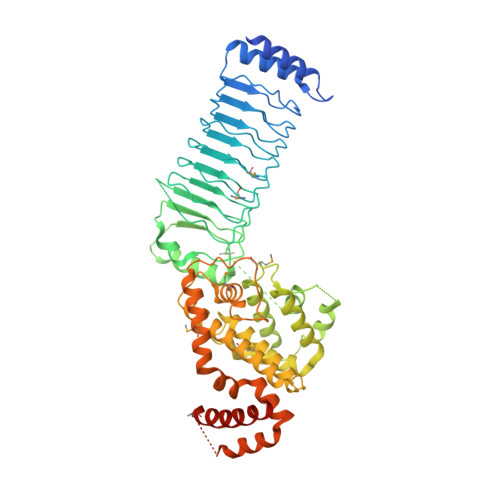
Structure of a Shigella effector reveals a new class of ubiquitin ligases
Zhu, Y., Li, H., Hu, L., Wang, J., Zhou, Y., Pang, Z., Liu, L., Shao, F.(2008) Nat Struct Mol Biol 15: 1302-1308
- PubMed: 18997779
- DOI: https://doi.org/10.1038/nsmb.1517
- Primary Citation of Related Structures:
3CVR - PubMed Abstract:
Bacterial pathogens have evolved effector proteins with ubiquitin E3 ligase activities through structural mimicking. Here we report the crystal structure of the Shigella flexneri type III effector IpaH3, a member of the leucine-rich repeat (LRR)-containing bacterial E3 family. The LRR domain is structurally similar to Yersinia pestis YopM and potentially binds to substrates. The structure of the C-terminal E3 domain differs from the typical RING- and HECT-type E3s. IpaH3 synthesizes a Lys48-linked ubiquitin chain, and the reaction requires noncovalent binding between ubiquitin and a specific E2, UbcH5. Free ubiquitin serves as an acceptor for IpaH3-catalyzed ubiquitin transfer. Cys363 within a conserved CXD motif acts as a nucleophile to catalyze ubiquitin transfer through a transthiolation reaction. The D365N mutant is devoid of E3 activities but turns into a potent ubiquitin-E2 thioesterase. Our analysis establishes a structurally and mechanistically distinct class of ubiquitin ligases found exclusively in pathogenic or symbiotic bacteria.
Organizational Affiliation:
National Institute of Biological Sciences, 7# Science Park Road, Zhongguancun Life Science Park, Beijing 102206, China.