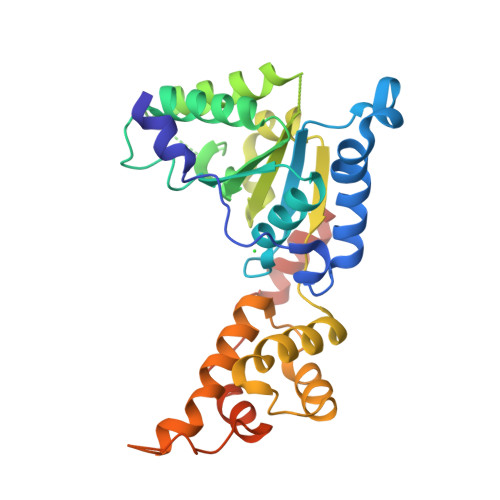
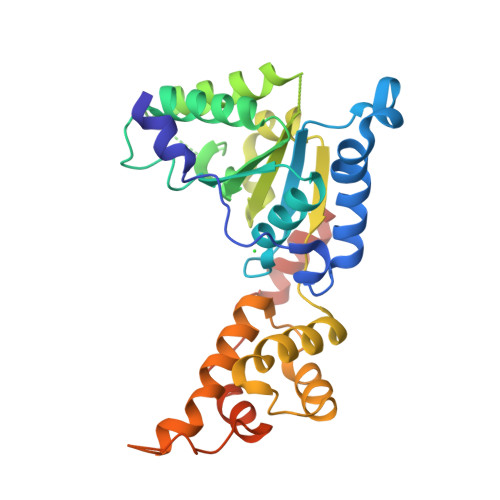
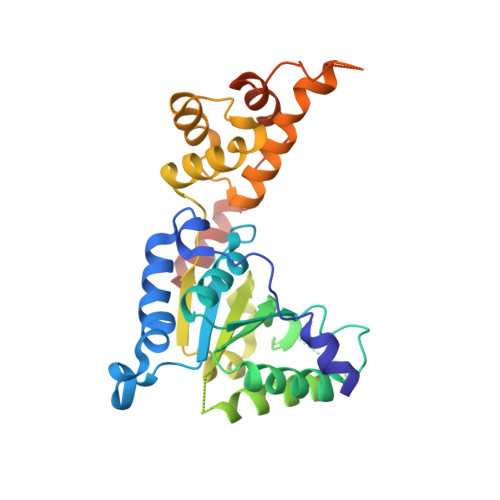
Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin.
Roll-Mecak, A., Vale, R.D.(2008) Nature 451: 363-367
- PubMed: 18202664
- DOI: https://doi.org/10.1038/nature06482
- Primary Citation of Related Structures:
3B9P - PubMed Abstract:
Spastin, the most common locus for mutations in hereditary spastic paraplegias, and katanin are related microtubule-severing AAA ATPases involved in constructing neuronal and non-centrosomal microtubule arrays and in segregating chromosomes. The mechanism by which spastin and katanin break and destabilize microtubules is unknown, in part owing to the lack of structural information on these enzymes. Here we report the X-ray crystal structure of the Drosophila spastin AAA domain and provide a model for the active spastin hexamer generated using small-angle X-ray scattering combined with atomic docking. The spastin hexamer forms a ring with a prominent central pore and six radiating arms that may dock onto the microtubule. Helices unique to the microtubule-severing AAA ATPases surround the entrances to the pore on either side of the ring, and three highly conserved loops line the pore lumen. Mutagenesis reveals essential roles for these structural elements in the severing reaction. Peptide and antibody inhibition experiments further show that spastin may dismantle microtubules by recognizing specific features in the carboxy-terminal tail of tubulin. Collectively, our data support a model in which spastin pulls the C terminus of tubulin through its central pore, generating a mechanical force that destabilizes tubulin-tubulin interactions within the microtubule lattice. Our work also provides insights into the structural defects in spastin that arise from mutations identified in hereditary spastic paraplegia patients.
Organizational Affiliation:
Howard Hughes Medical Institute and Department of Cellular and Molecular Pharmacology, University of California, San Francisco, 600 16th Street, San Francisco, California 94158, USA.