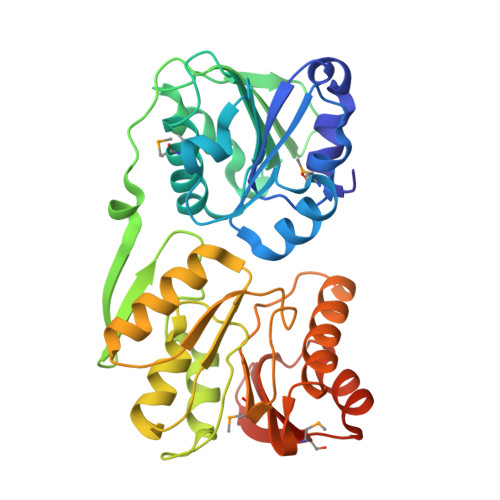
Functional specialization of domains tandemly duplicated within 16S rRNA methyltransferase RsmC
Sunita, S., Purta, E., Durawa, M., Tkaczuk, K.L., Swaathi, J., Bujnicki, J.M., Sivaraman, J.(2007) Nucleic Acids Res 35: 4264-4274
- PubMed: 17576679
- DOI: https://doi.org/10.1093/nar/gkm411
- Primary Citation of Related Structures:
2PJD - PubMed Abstract:
RNA methyltransferases (MTases) are important players in the biogenesis and regulation of the ribosome, the cellular machine for protein synthesis. RsmC is a MTase that catalyzes the transfer of a methyl group from S-adenosyl-l-methionine (SAM) to G1207 of 16S rRNA. Mutations of G1207 have dominant lethal phenotypes in Escherichia coli, underscoring the significance of this modified nucleotide for ribosome function. Here we report the crystal structure of E. coli RsmC refined to 2.1 A resolution, which reveals two homologous domains tandemly duplicated within a single polypeptide. We characterized the function of the individual domains and identified key residues involved in binding of rRNA and SAM, and in catalysis. We also discovered that one of the domains is important for the folding of the other. Domain duplication and subfunctionalization by complementary degeneration of redundant functions (in particular substrate binding versus catalysis) has been reported for many enzymes, including those involved in RNA metabolism. Thus, RsmC can be regarded as a model system for functional streamlining of domains accompanied by the development of dependencies concerning folding and stability.
Organizational Affiliation:
Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543.