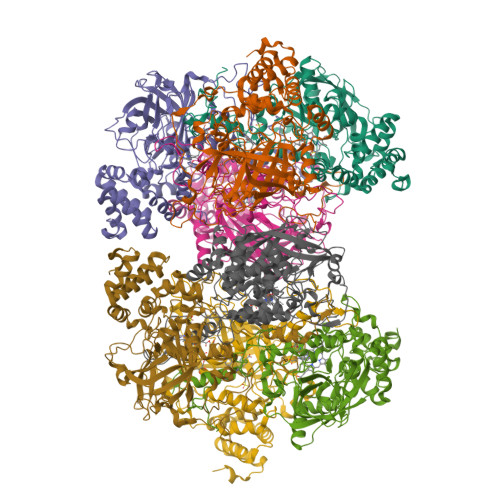
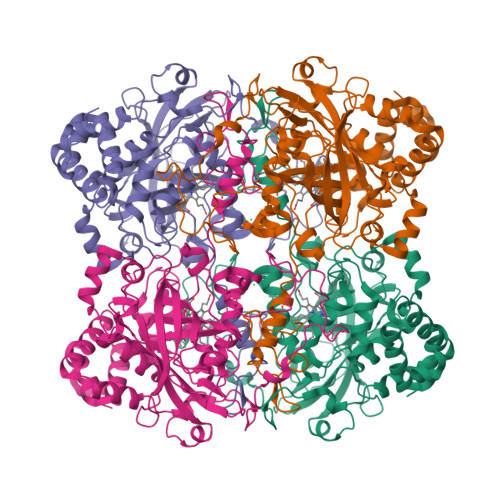
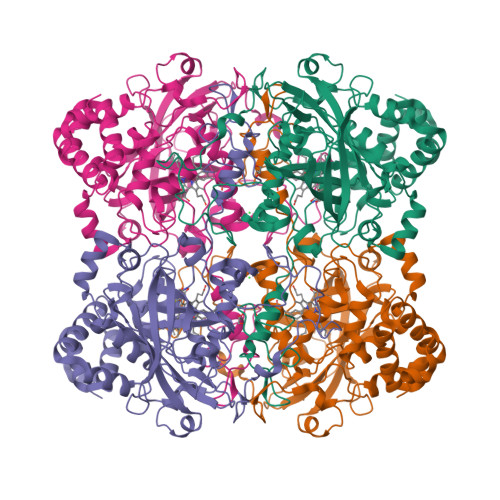
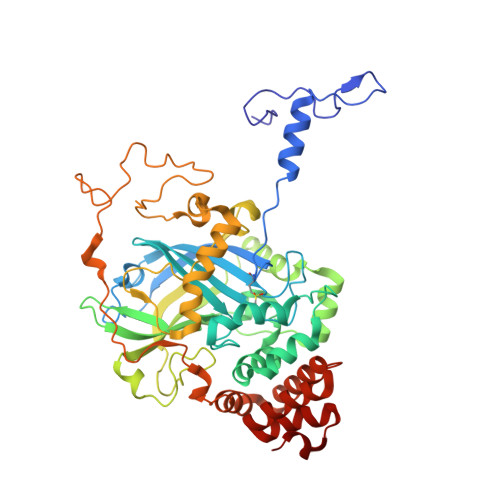
The first structure of a cold-active catalase from Vibrio salmonicida at 1.96A reveals structural aspects of cold adaptation
Riise, E.K., Lorentzen, M.S., Helland, R., Smalas, A.O., Leiros, H.K.S., Willassen, N.P.(2007) Acta Crystallogr D Biol Crystallogr 63: 135-148
- PubMed: 17242507
- DOI: https://doi.org/10.1107/S0907444906043812
- Primary Citation of Related Structures:
2ISA - PubMed Abstract:
The cold-adapted catalase from the fish-pathogenic bacterium Vibrio salmonicida (VSC) has recently been characterized and shown to be two times more catalytically efficient compared with catalase from the mesophilic human pathogen Proteus mirabilis [PMC; Lorentzen et al. (2006), Extremophiles, 10, 427-440]. VSC is also less temperature-stable, with a half-life of 5 min at 333 K compared with 50 min for PMC. This was the background for solving the crystal structure of the cold-adapted VSC to 1.96 A and performing an extensive structural comparison of VSC and PMC. The comparison revealed that the entrance (the major channel) leading to the catalytically essential haem group, is locally more flexible and slightly wider in VSC. This might explain the enhanced catalytic efficiency of the nearly diffusion-controlled degradation of hydrogen peroxide into water and molecular oxygen in VSC. The reduced thermal stability of the cold-adapted VSC may be explained by a reduced number of ion-pair networks. The four C-terminal alpha-helices are displaced in the structures, probably owing to missing ionic interactions in VSC compared with PMC, and this is postulated as an initiation site for unfolding the cold-adapted enzyme. VSC is the first crystal structure reported of a cold-adapted monofunctional haem-containing catalase.
Organizational Affiliation:
The Norwegian Structural Biology Centre (NorStruct), Department of Chemistry, Faculty of Science, University of Tromsø, N-9037 Tromsø, Norway.