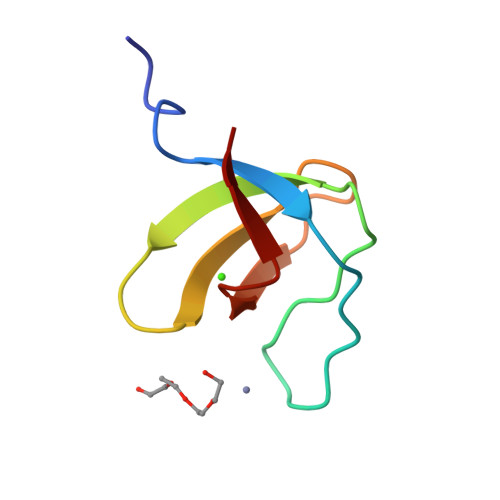
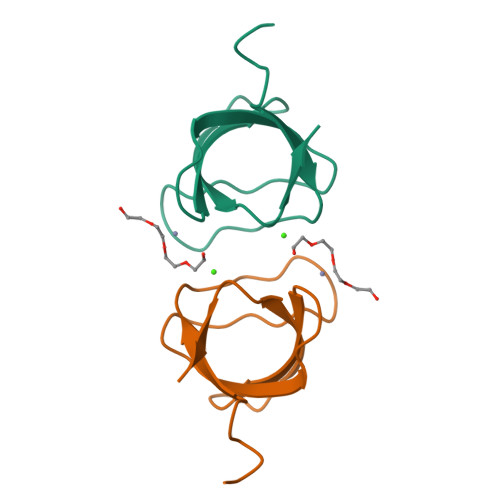
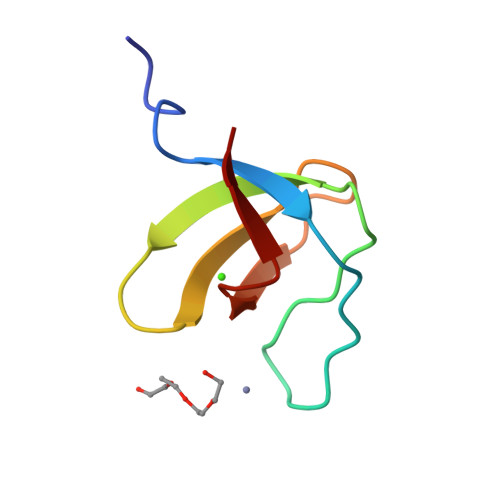
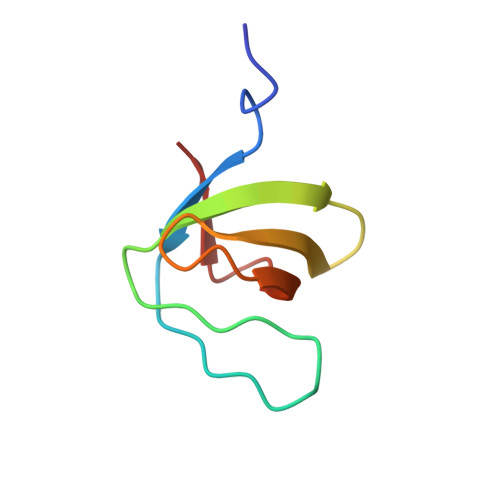
Crystal structure analysis and solution studies of human Lck-SH3; zinc-induced homodimerization competes with the binding of proline-rich motifs.
Romir, J., Lilie, H., Egerer-Sieber, C., Bauer, F., Sticht, H., Muller, Y.A.(2007) J Mol Biology 365: 1417-1428
- PubMed: 17118402
- DOI: https://doi.org/10.1016/j.jmb.2006.10.058
- Primary Citation of Related Structures:
2IIM - PubMed Abstract:
In cytosolic Src-type tyrosine kinases the Src-type homology 3 (SH3) domain binds to an internal proline-rich motif and the presence or the absence of this interaction modulates the kinase enzymatic activity. The Src-type kinase Lck plays an important role during T-cell activation and development, since it phosphorylates the T-cell antigen receptor in an early step of the activation pathway. We have determined the crystal structure of the SH3 domain from Lck kinase at a near-atomic resolution of 1.0 A. Unexpectedly, the Lck-SH3 domain forms a symmetrical homodimer in the crystal and the dimer comprises two identical zinc-binding sites in the interface. The atomic interactions formed across the dimer interface resemble strikingly those observed between SH3 domains and their canonical proline-rich ligands, since almost identical residues participate in both contacts. Ultracentrifugation experiments confirm that in the presence of zinc ions, the Lck-SH3 domain also forms dimers in solution. The Zn(2+) dissociation constant from the Lck-SH3 dimer is estimated to be lower than 100 nM. Moreover, upon addition of a proline-rich peptide with a sequence corresponding to the recognition segment of the herpesviral regulatory protein Tip, competition between zinc-induced homodimerization and binding of the peptide can be detected by both fluorescence spectroscopy and analytical ultracentrifugation. These results suggest that in vivo, too, competition between Lck-SH3 homodimerization and binding of regulatory proline-rich sequence motifs possibly represents a novel mechanism by which kinase activity is modulated. Because the residues that form the zinc-binding site are highly conserved among Lck orthologues but not in other Src-type kinases, the mechanism might be peculiar to Lck and to its role in the initial steps of T-cell activation.
Organizational Affiliation:
Lehrstuhl für Biotechnik, Institut für Biologie, Friedrich-Alexander-Universität Erlangen-Nürnberg, Germany.