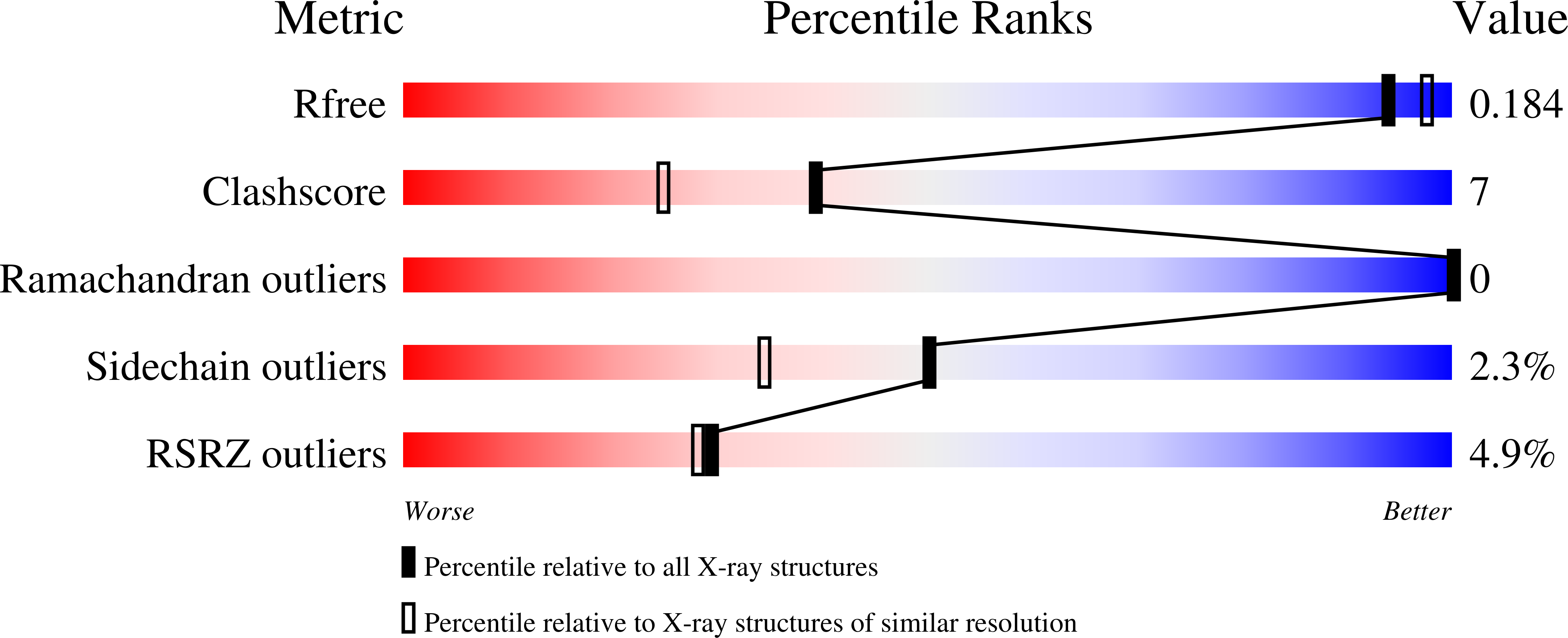
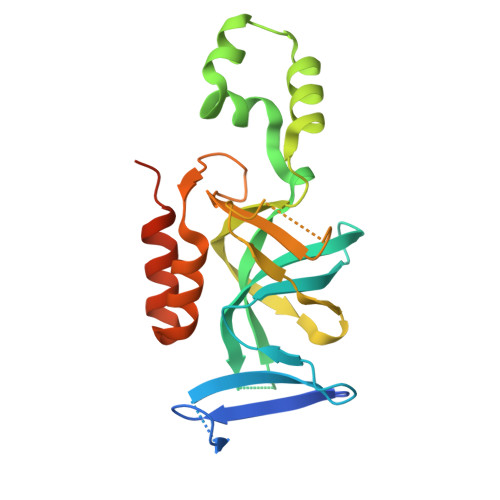
Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 A resolution.
Hou, Z., Danzer, J.R., Fox, C.A., Keck, J.L.(2006) Protein Sci 15: 1182-1186
- PubMed: 16641491
- DOI: https://doi.org/10.1110/ps.052061006
- Primary Citation of Related Structures:
2FL7 - PubMed Abstract:
Sir3p is a silent-information-regulator (SIR) protein required for the assembly of a transcriptionally "silent" chromatin structure at telomeres and the cryptic HM mating-type loci in Saccharomyces cerevisiae. Sir3p contains a putative "bromo adjacent homology" (BAH) domain at its N terminus that shares strong sequence similarity with the BAH domain of a subunit of the origin recognition complex (ORC), Orc1p. The Orc1p-BAH domain forms a well-defined complex with the ORC interaction region (OIR) of another Sir protein, Sir1p, which targets formation of silent chromatin to the HM-loci. Interestingly, despite sequence similarity of the Sir3p and Orc1p BAH domains and Sir3p's established importance in silencing, Sir3p does not bind the Sir1p-OIR. Here we report the 1.95 A resolution crystal structure of the Sir3p-BAH domain. The structure reveals two key features that can account for Sir3p-BAH domain's inability to interact with Sir1p. First, several Orc1p-BAH domain residues known to directly contact Sir1p are altered in the Sir3p-BAH domain. Second, a critical OIR-binding pocket present on the surface of the Orc1p-BAH domain is "filled" in the Sir3p-BAH domain structure, potentially making it inaccessible to Sir1p. These findings imply that the Sir3p-BAH domain structure has evolved for functions distinct from those of the Orc1p-BAH domain.
Organizational Affiliation:
Department of Biomolecular Chemistry, University of Wisconsin School of Medicine and Public Health, Madison, Wisconsin 53706-1532, USA.