The First Holocomplex Structure of Ribonucleotide Reductase Gives New Insight Into its Mechanism of Action
Uppsten, M., Farnegardh, M., Domkin, V., Uhlin, U.(2006) J Mol Biol 359: 365
- PubMed: 16631785
- DOI: https://doi.org/10.1016/j.jmb.2006.03.035
- Primary Citation of Related Structures:
2BQ1 - PubMed Abstract:
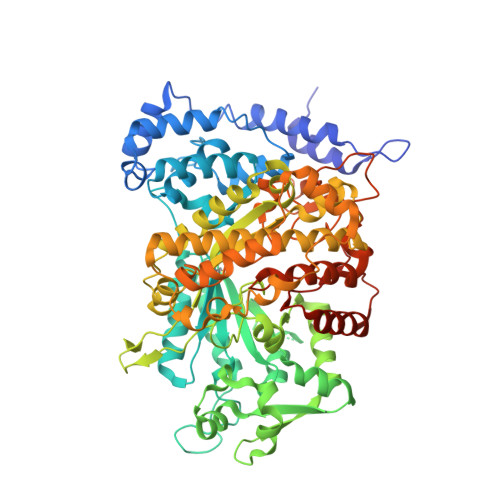
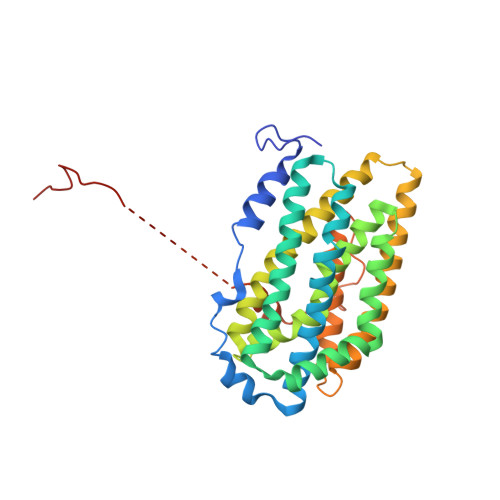
Ribonucleotide reductase is an indispensable enzyme for all cells, since it catalyses the biosynthesis of the precursors necessary for both building and repairing DNA. The ribonucleotide reductase class I enzymes, present in all mammals as well as in many prokaryotes and DNA viruses, are composed mostly of two homodimeric proteins, R1 and R2. The reaction involves long-range radical transfer between the two proteins. Here, we present the first crystal structure of a ribonucleotide reductase R1/R2 holocomplex. The biological relevance of this complex is based on the binding of the R2 C terminus in the hydrophobic cleft of R1, an interaction proven to be crucial for enzyme activity, and by the fact that all conserved amino acid residues in R2 are facing the R1 active sites. We suggest that the asymmetric R1/R2 complex observed in the 4A crystal structure of Salmonella typhimurium ribonucleotide reductase represents an intermediate stage in the reaction cycle, and at the moment of reaction the homodimers transiently form a tight symmetric complex.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Uppsala Biomedical Center, Box 590, SE-751 24 Uppsala, Sweden.