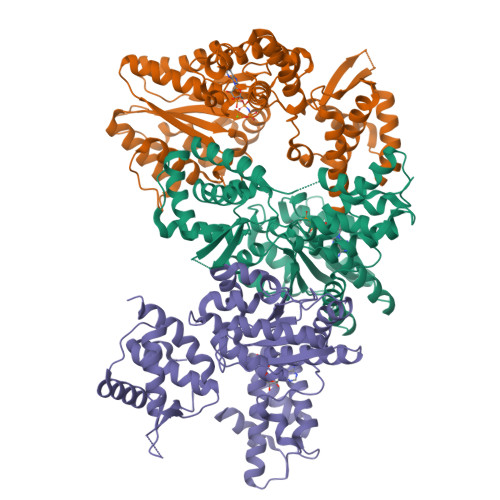
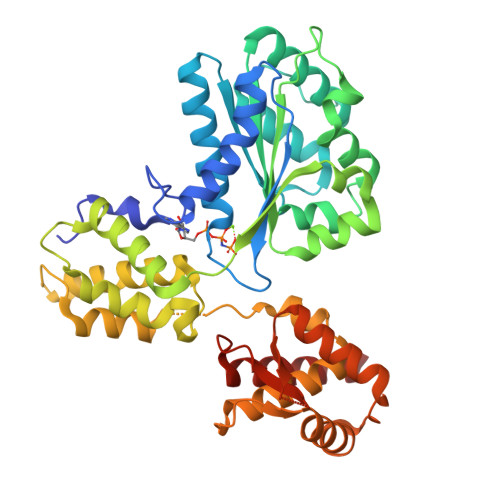
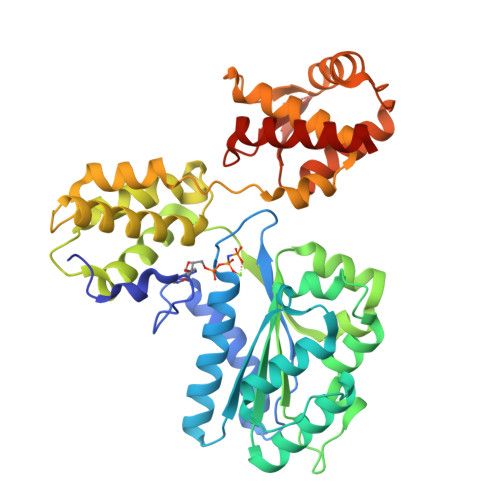
Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
Singleton, M.R., Morales, R., Grainge, I., Cook, N., Isupov, M.N., Wigley, D.B.(2004) J Mol Biology 343: 547
- PubMed: 15465044
- DOI: https://doi.org/10.1016/j.jmb.2004.08.044
- Primary Citation of Related Structures:
1W5S, 1W5T - PubMed Abstract:
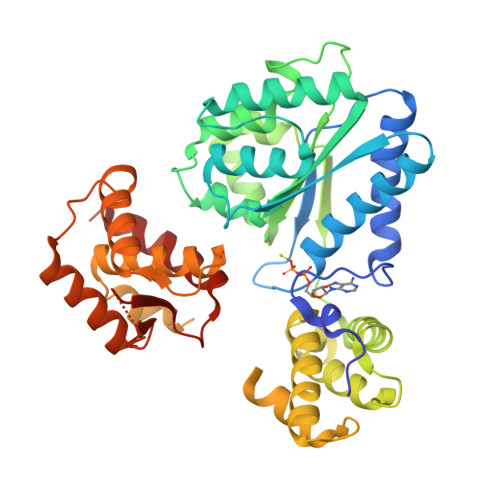
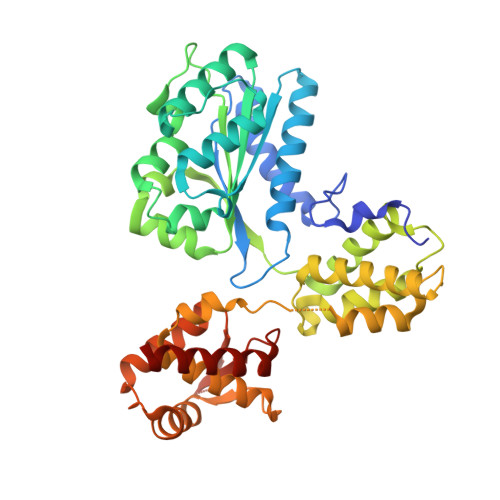
Archaea contain one or more proteins with homology to eukaryotic ORC/Cdc6 proteins. Sequence analysis suggests the existence of at least two subfamilies of these proteins, for which we propose the nomenclature ORC1 and ORC2. We have determined crystal structures of the ORC2 protein from the archaeon Aeropyrum pernix in complexes with ADP or a non-hydrolysable ATP analogue, ADPNP. Between two crystal forms, there are three crystallographically independent views of the ADP complex and two of the ADPNP complex. The protein molecules in the three complexes with ADP adopt very different conformations, while the two complexes with ADPNP are the same. These structures indicate that there is considerable conformational flexibility in ORC2 but that ATP binding stabilises a single conformation. We show that the ORC2 protein can bind DNA, and that this activity is associated with the C-terminal domain of the protein. We present a model for the interaction of the winged helix (WH) domain of ORC2 with DNA that differs from that proposed previously for Pyrobaculum aerophilum ORC/Cdc6.
Organizational Affiliation:
Cancer Research UK Clare Hall Laboratories, The London Research Institute, Blanche Lane, South Mimms, Potters Bar, Herts EN6 3LD, UK.