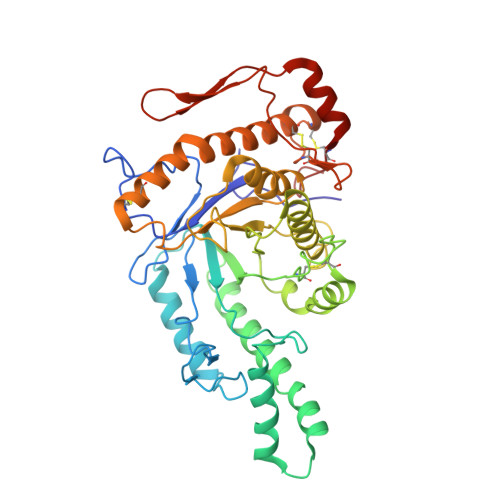
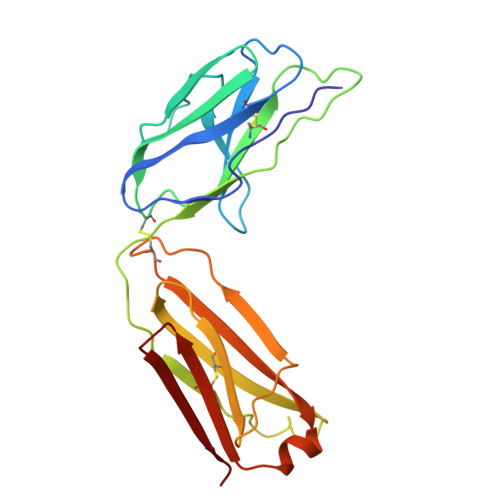
Cryo-EM Structure of Human Hyaluronidase PH-20.
Im, S.B., Song, H.N., Jeong, T.K., Kim, N., Kim, K., Park, S.J., Oh, B.H.(2024) Proteins
- PubMed: 39722545
- DOI: https://doi.org/10.1002/prot.26788
- Primary Citation of Related Structures:
9JUB - PubMed Abstract:
PH-20 is a specific type of hyaluronidase that plays a critical role in the fertilization process by facilitating the initial binding of sperm to the glycoprotein layer surrounding the oocyte and subsequently breaking down hyaluronic acid polymers in the cumulus cell layer. PH-20 contains an epidermal growth factor (EGF)-like domain, which may be involved in the recognition of the glycoprotein layer in addition to the catalytic domain. Herein, we report the structure of human PH-20 determined by cryogenic electron microscopy. Comparative analyses of the PH-20 structure with two other available hyaluronidase structures reveal a general similarity in the central catalytic domains, including the conservation of catalytically essential residues at the equivalent spatial positions. However, unique difference is found in the EGF-like domain, characterized by a longer sequence that is likely to form a flexibly anchored β-hairpin containing a disulfide bond.
Organizational Affiliation:
Department of Biological Sciences, KAIST Institute for the Biocentury, Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea.