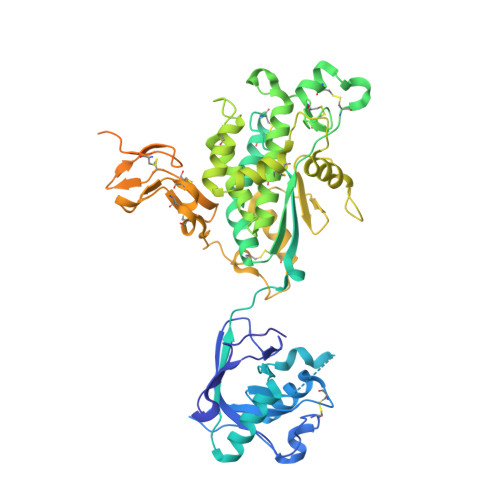
Functional implication of the homotrimeric multidomain vacuolar sorting receptor 1 (VSR1) from Arabidopsis thaliana.
Park, H., Youn, B., Park, D.J., Puthanveettil, S.V., Kang, C.(2024) Sci Rep 14: 9622-9622
- PubMed: 38671060
- DOI: https://doi.org/10.1038/s41598-024-57975-2
- Primary Citation of Related Structures:
9B1R - PubMed Abstract:
The vacuolar sorting receptors (VSRs) are specific to plants and are responsible for sorting and transporting particular proteins from the trans-Golgi network to the vacuole. This process is critically important for various cellular functions, including storing nutrients during seed development. Despite many years of intense studies on VSRs, a complete relation between function and structure has not yet been revealed. Here, we present the crystal structure of the entire luminal region of glycosylated VSR1 from Arabidopsis thaliana (AtVSR1) for the first time. The structure provides insights into the tertiary and quaternary structures of VSR1, which are composed of an N-terminal protease-associated (PA) domain, a unique central region, and one epidermal growth factor (EGF)-like domain followed by two disordered EGF-like domains. The structure of VSR1 exhibits unique characteristics, the significance of which is yet to be fully understood.
Organizational Affiliation:
X-Ray Core, UF Scripps Biomedical Research, University of Florida, Jupiter, FL, 33458, USA.