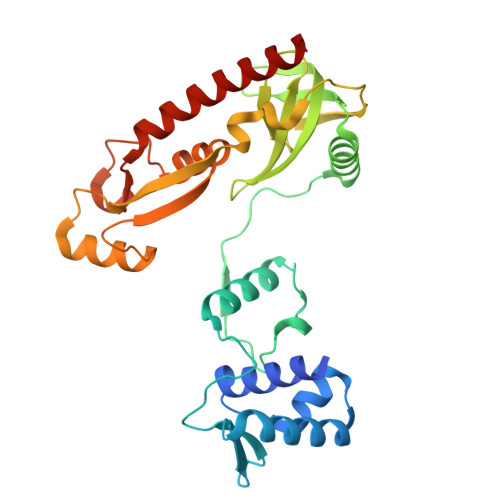
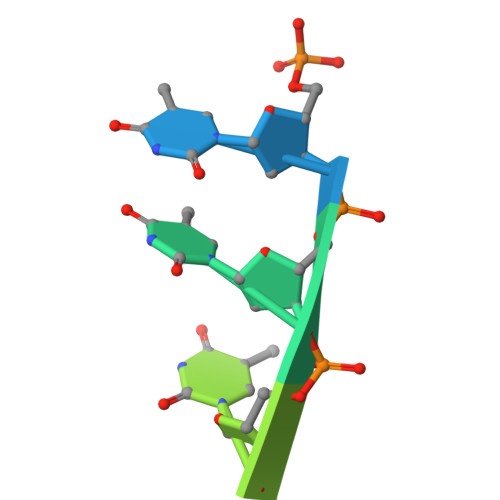
Bacterial WYL domain transcriptional repressors sense single-stranded DNA to control gene expression.
Blankenchip, C.L., Corbett, K.D.(2024) Nucleic Acids Res
- PubMed: 39588753
- DOI: https://doi.org/10.1093/nar/gkae1101
- Primary Citation of Related Structures:
9C5G - PubMed Abstract:
Bacteria encode a wide array of immune systems to protect themselves against ubiquitous bacteriophages and foreign DNA elements. While these systems' molecular mechanisms are becoming increasingly well known, their regulation remains poorly understood. Here, we show that an immune system-associated transcriptional repressor of the wHTH-WYL-WCX family, CapW, directly binds single-stranded DNA to sense DNA damage and activate expression of its associated immune system. We show that CapW mediates increased expression of a reporter gene in response to DNA damage in a host cell. CapW directly binds single-stranded DNA by-products of DNA repair through its WYL domain, causing a conformational change that releases the protein from double-stranded DNA. In an Escherichia coli CBASS system with an integrated capW gene, we find that CapW-mediated transcriptional activation is important for this system's ability to prevent induction of a λ prophage. Overall, our data reveal the molecular mechanisms of WYL-domain transcriptional repressors, and provide an example of how bacteria can balance the protective benefits of carrying anti-phage immune systems against the inherent risk of these systems' aberrant activation.
Organizational Affiliation:
Biomedical Sciences Graduate Program, University of California, San Diego, 9500 Gilman Dr. La Jolla, CA 92093, USA.