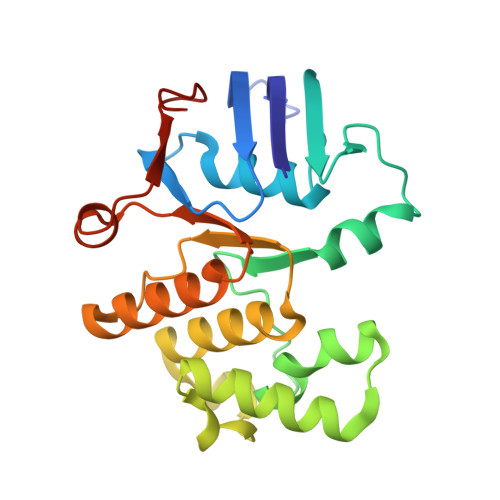
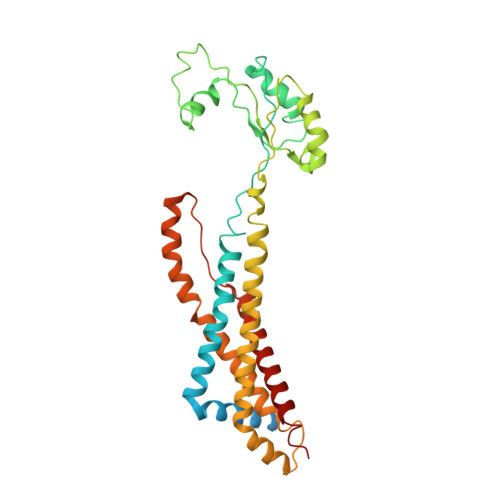
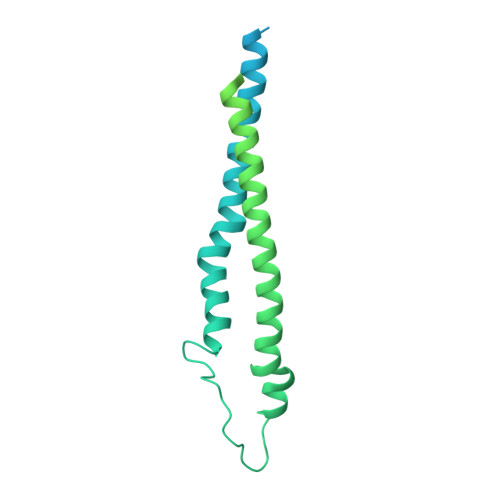
Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Chen, Y., Gu, J., Yang, B., Yang, L., Pang, J., Luo, Q., Li, Y., Li, D., Deng, Z., Dong, C., Dong, H., Zhang, Z.(2024) PLoS Biol 22: e3002628-e3002628
- PubMed: 38814940
- DOI: https://doi.org/10.1371/journal.pbio.3002628
- Primary Citation of Related Structures:
8X61, 8Y3X - PubMed Abstract:
The peptidoglycan (PG) layer is a critical component of the bacterial cell wall and serves as an important target for antibiotics in both gram-negative and gram-positive bacteria. The hydrolysis of septal PG (sPG) is a crucial step of bacterial cell division, facilitated by FtsEX through an amidase activation system. In this study, we present the cryo-EM structures of Escherichia coli FtsEX and FtsEX-EnvC in the ATP-bound state at resolutions of 3.05 Å and 3.11 Å, respectively. Our PG degradation assays in E. coli reveal that the ATP-bound conformation of FtsEX activates sPG hydrolysis of EnvC-AmiB, whereas EnvC-AmiB alone exhibits autoinhibition. Structural analyses indicate that ATP binding induces conformational changes in FtsEX-EnvC, leading to significant differences from the apo state. Furthermore, PG degradation assays of AmiB mutants confirm that the regulation of AmiB by FtsEX-EnvC is achieved through the interaction between EnvC-AmiB. These findings not only provide structural insight into the mechanism of sPG hydrolysis and bacterial cell division, but also have implications for the development of novel therapeutics targeting drug-resistant bacteria.
Organizational Affiliation:
Department of Clinical Laboratory, Zhongnan Hospital of Wuhan University, School of Pharmaceutical Sciences, Wuhan University, Wuhan, China.