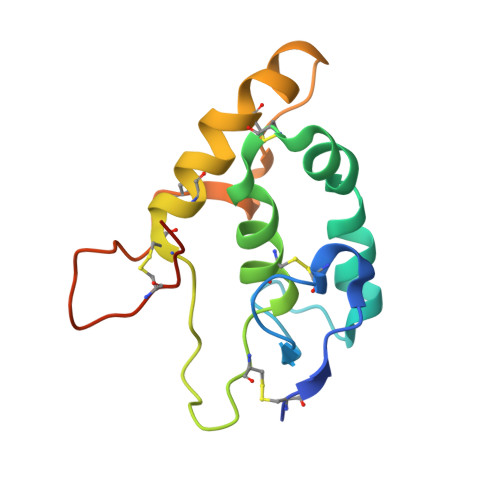
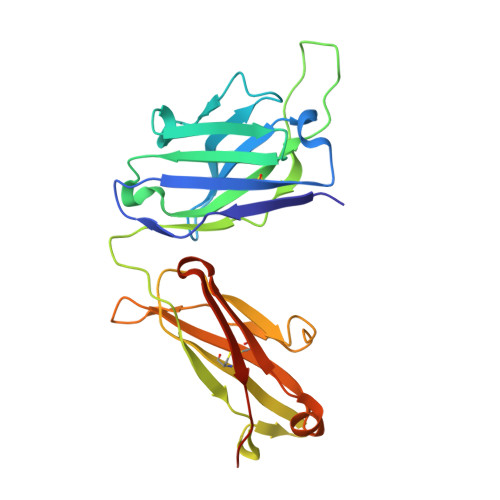
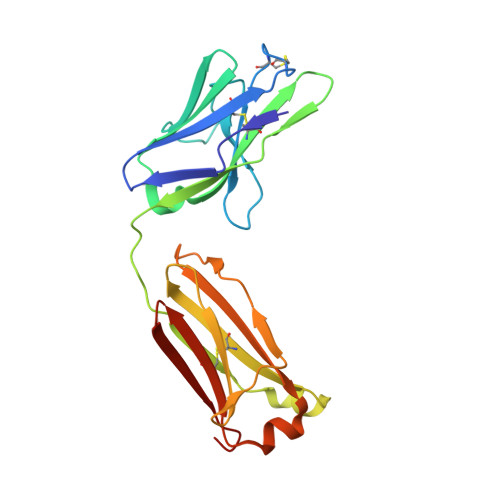
Identification and functional validation of FZD8-specific antibodies.
Li, N., Ge, Q., Guo, Q., Tao, Y.(2023) Int J Biol Macromol 254: 127846
- PubMed: 37926311
- DOI: https://doi.org/10.1016/j.ijbiomac.2023.127846
- Primary Citation of Related Structures:
8X0T - PubMed Abstract:
The Wnt pathway is an evolutionarily conserved pathway involved in stem cell homeostasis and tissue regeneration. Aberrant signaling in the Wnt pathway is highly associated with cancer. Developing antibodies to block overactivation of Frizzled receptors (FZDs), the main receptors in the Wnt pathway, is one of the viable options for treating cancer. However, obtaining isoform-specific antibodies is often challenging due to the high degree of homology among the ten FZDs. In this study, by using a synthetic library, we identified an antibody named pF8_AC3 that preferentially binds to FZD8. Guided by the structure of the complex of pF8_AC3 and FZD8, a second-generation targeted library was further constructed, and finally, the FZD8-specific antibody sF8_AG6 was obtained. Cell-based assays showed that these antibodies could selectively block FZD8-mediated signaling activation. Taken together, these antibodies have the potential to be developed into therapeutic drugs in the future.
Organizational Affiliation:
MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Center for Advanced Interdisciplinary Science and Biomedicine of IHM, Hefei National Research Center for Physical Sciences at the Microscale, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China; Biomedical Sciences and Health Laboratory of Anhui Province, University of Science and Technology of China, Hefei, China.