Discovery of (4-Pyrazolyl)-2-aminopyrimidines as Potent and Selective Inhibitors of Cyclin-Dependent Kinase 2.
Hummel, J.R., Xiao, K.J., Yang, J.C., Epling, L.B., Mukai, K., Ye, Q., Xu, M., Qian, D., Huo, L., Weber, M., Roman, V., Lo, Y., Drake, K., Stump, K., Covington, M., Kapilashrami, K., Zhang, G., Ye, M., Diamond, S., Yeleswaram, S., Macarron, R., Deller, M.C., Wee, S., Kim, S., Wang, X., Wu, L., Yao, W.(2024) J Med Chem 67: 3112-3126
- PubMed: 38325398
- DOI: https://doi.org/10.1021/acs.jmedchem.3c02287
- Primary Citation of Related Structures:
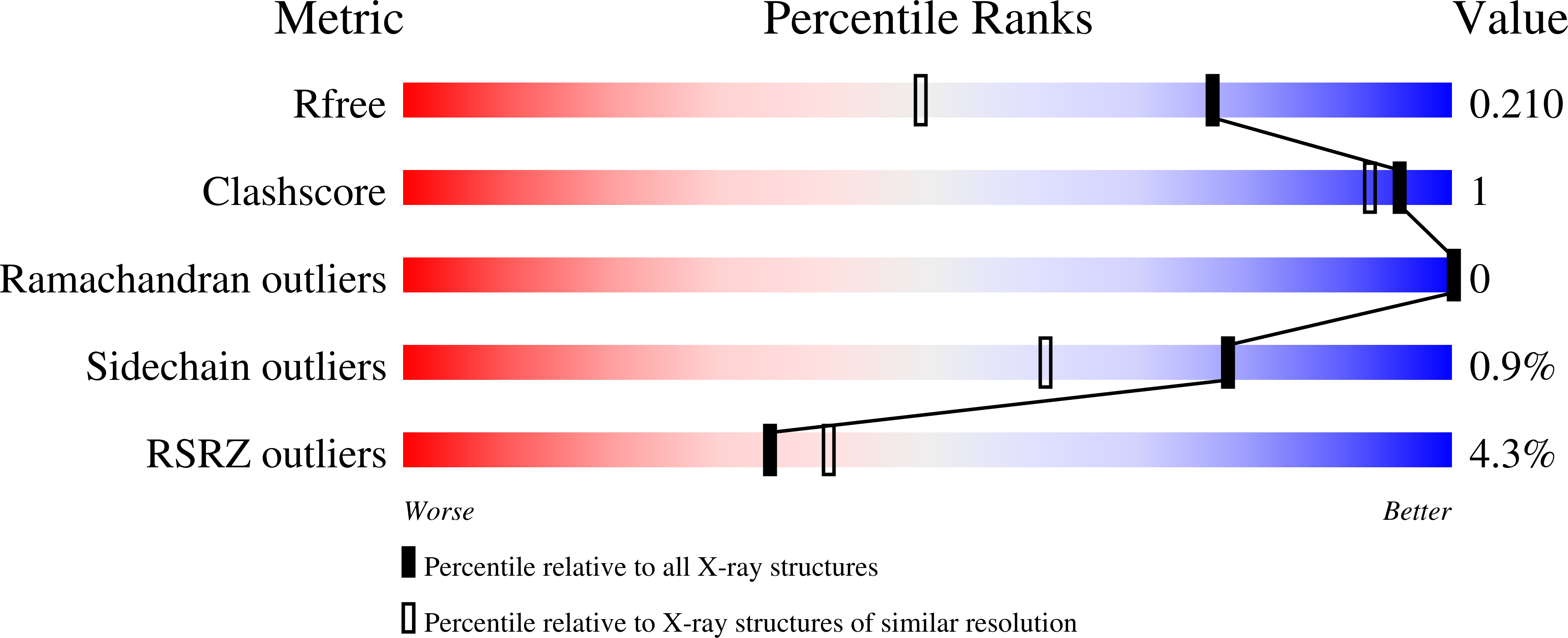
8UV0 - PubMed Abstract:
CDK2 is a critical regulator of the cell cycle. For a variety of human cancers, the dysregulation of CDK2/cyclin E1 can lead to tumor growth and proliferation. Historically, early efforts to develop CDK2 inhibitors with clinical applications proved unsuccessful due to challenges in achieving selectivity over off-target CDK isoforms with associated toxicity. In this report, we describe the discovery of (4-pyrazolyl)-2-aminopyrimidines as a potent class of CDK2 inhibitors that display selectivity over CDKs 1, 4, 6, 7, and 9. SAR studies led to the identification of compound 17 , a kinase selective and highly potent CDK2 inhibitor (IC 50 = 0.29 nM). The evaluation of 17 in CCNE1 -amplified mouse models shows the pharmacodynamic inhibition of CDK2, measured by reduced Rb phosphorylation, and antitumor activity.
Organizational Affiliation:
Incyte Research Institute, Incyte Corporation, 1801 Augustine Cut-Off, Wilmington, Delaware 19803, United States.