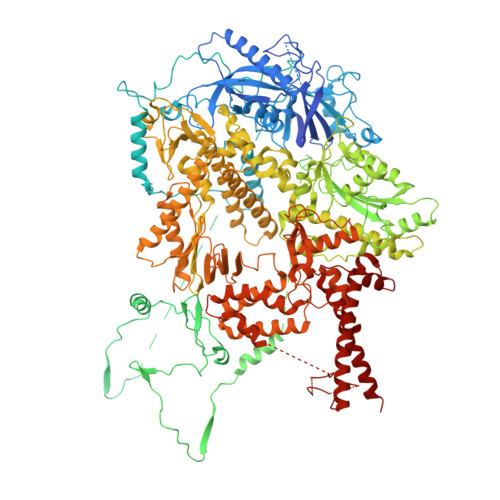
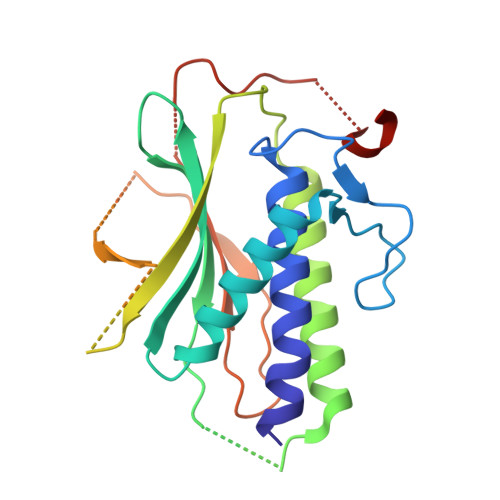
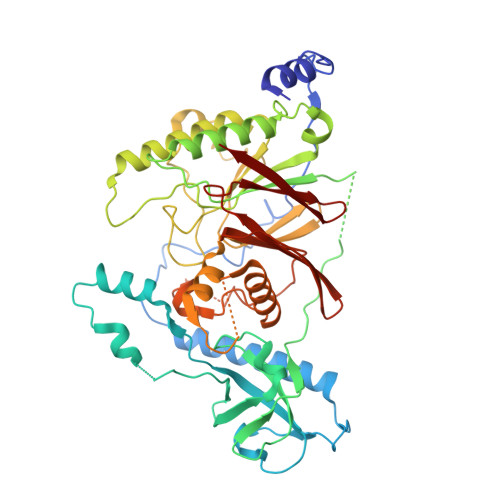
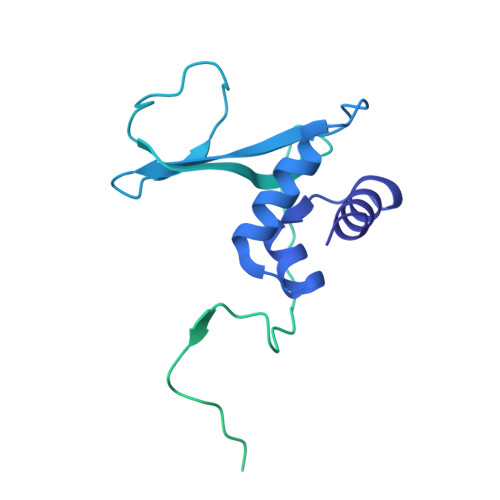
Cryo-EM structure of the Rev1-Pol zeta holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis.
Malik, R., Johnson, R.E., Ubarretxena-Belandia, I., Prakash, L., Prakash, S., Aggarwal, A.K.(2024) Nat Struct Mol Biol 31: 1394-1403
- PubMed: 38720088
- DOI: https://doi.org/10.1038/s41594-024-01302-w
- Primary Citation of Related Structures:
8TLQ, 8TLT - PubMed Abstract:
Rev1-Polζ-dependent translesion synthesis (TLS) of DNA is crucial for maintaining genome integrity. To elucidate the mechanism by which the two polymerases cooperate in TLS, we determined the cryogenic electron microscopic structure of the Saccharomyces cerevisiae Rev1-Polζ holocomplex in the act of DNA synthesis (3.53 Å). We discovered that a composite N-helix-BRCT module in Rev1 is the keystone of Rev1-Polζ cooperativity, interacting directly with the DNA template-primer and with the Rev3 catalytic subunit of Polζ. The module is positioned akin to the polymerase-associated domain in Y-family TLS polymerases and is set ideally to interact with PCNA. We delineate the full extent of interactions that the carboxy-terminal domain of Rev1 makes with Polζ and identify potential new druggable sites to suppress chemoresistance from first-line chemotherapeutics. Collectively, our results provide fundamental new insights into the mechanism of cooperativity between Rev1 and Polζ in TLS.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA. radhika.malik@mssm.edu.