Targeting bacterial nickel transport with aspergillomarasmine A suppresses virulence-associated Ni-dependent enzymes.
Sychantha, D., Chen, X., Koteva, K., Prehna, G., Wright, G.D.(2024) Nat Commun 15: 4036-4036
- PubMed: 38740750
- DOI: https://doi.org/10.1038/s41467-024-48232-1
- Primary Citation of Related Structures:
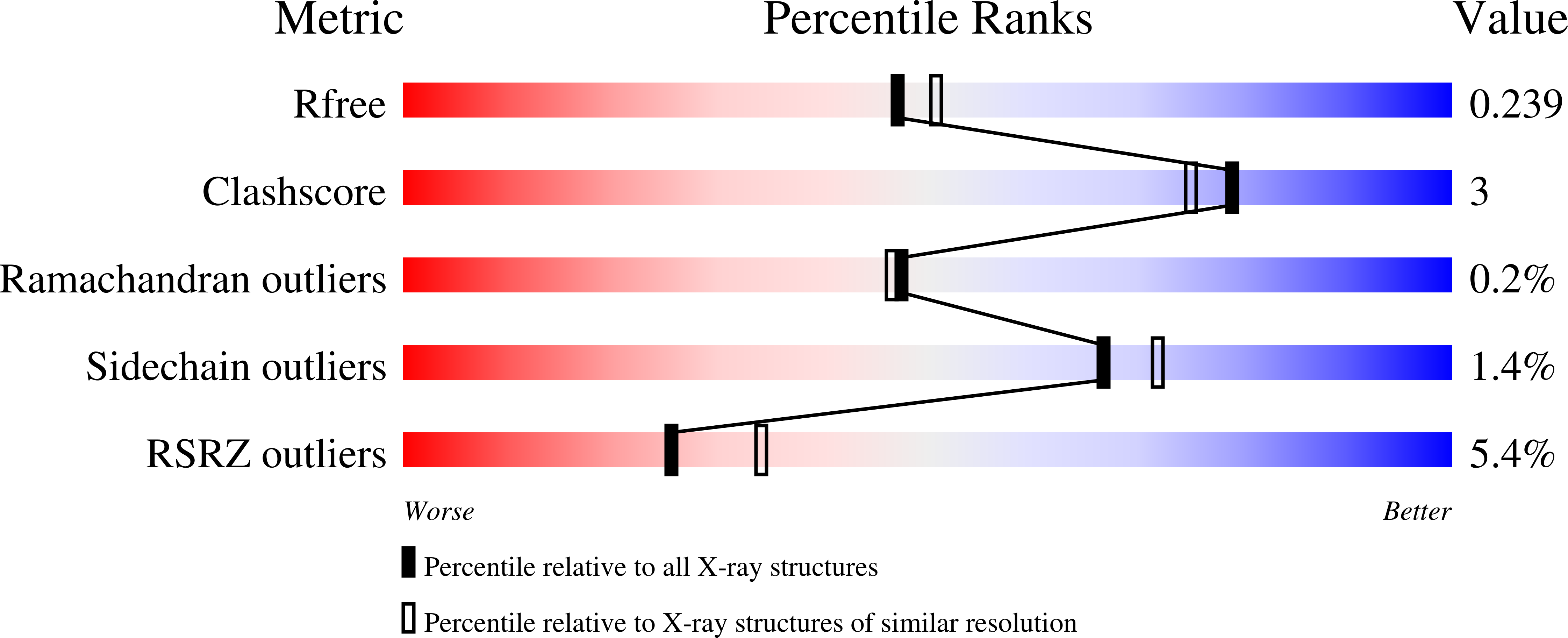
8SPM - PubMed Abstract:
Microbial Ni 2+ homeostasis underpins the virulence of several clinical pathogens. Ni 2+ is an essential cofactor in urease and [NiFe]-hydrogenases involved in colonization and persistence. Many microbes produce metallophores to sequester metals necessary for their metabolism and starve competing neighboring organisms. The fungal metallophore aspergillomarasmine A (AMA) shows narrow specificity for Zn 2+ , Ni 2+ , and Co 2+ . Here, we show that this specificity allows AMA to block the uptake of Ni 2+ and attenuate bacterial Ni-dependent enzymes, offering a potential strategy for reducing virulence. Bacterial exposure to AMA perturbs H 2 metabolism, ureolysis, struvite crystallization, and biofilm formation and shows efficacy in a Galleria mellonella animal infection model. The inhibition of Ni-dependent enzymes was aided by Zn 2+, which complexes with AMA and competes with the native nickelophore for the uptake of Ni 2+ . Biochemical analyses demonstrated high-affinity binding of AMA-metal complexes to NikA, the periplasmic substrate-binding protein of the Ni 2+ uptake system. Structural examination of NikA in complex with Ni-AMA revealed that the coordination geometry of Ni-AMA mimics the native ligand, Ni-(L-His) 2 , providing a structural basis for binding AMA-metal complexes. Structure-activity relationship studies of AMA identified regions of the molecule that improve NikA affinity and offer potential routes for further developing this compound as an anti-virulence agent.
Organizational Affiliation:
David Braley Centre for Antibiotic Discovery, McMaster University, Hamilton, ON, Canada.