Structural basis for recognition of the FLAG-tag by anti-FLAG M2.
Wouter Beugelink, J., Sweep, E., Janssen, B.J.C., Snijder, J., Pronker, M.F.(2024) J Mol Biol : 168649-168649
- PubMed: 38852931
- DOI: https://doi.org/10.1016/j.jmb.2024.168649
- Primary Citation of Related Structures:
8RMO - PubMed Abstract:
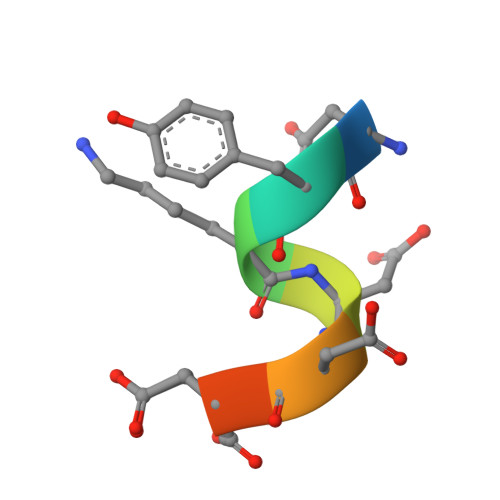
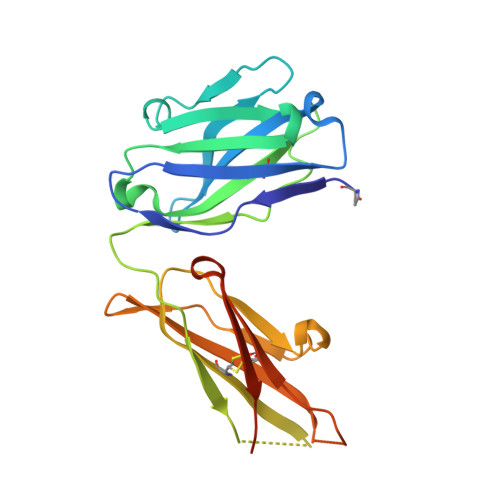
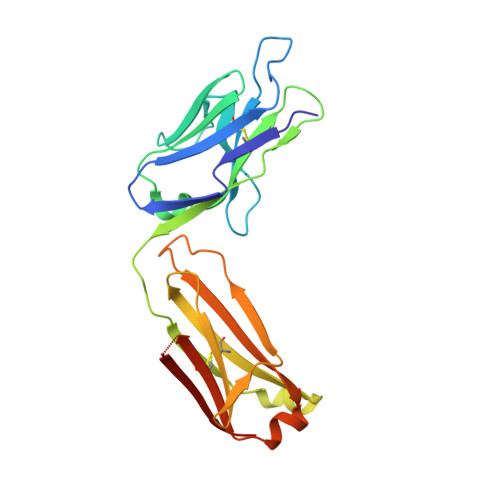
The FLAG-tag/anti-FLAG system is a widely used biochemical tool for protein detection and purification. Anti-FLAG M2 is the most popular antibody against the FLAG-tag, due to its ease of use, versatility, and availability in pure form or as bead conjugate. M2 binds N-terminal, C-terminal and internal FLAG-tags and binding is calcium-independent, but the molecular basis for the FLAG-tag specificity and recognition remains unresolved. Here we present an atomic resolution (1.17 Å) structure of the FLAG peptide in complex with the Fab of anti-FLAG M2, revealing key binding determinants. Five of the eight FLAG peptide residues form direct interactions with paratope residues. The FLAG peptide adopts a 3 10 helix conformation in complex with the Fab. These structural insights allowed us to rationally introduce point mutations on both the peptide and antibody side. We tested these by surface plasmon resonance, leading us to propose a shorter yet equally binding version of the FLAG-tag for the M2 antibody.
Organizational Affiliation:
Structural Biochemistry, Bijvoet Centre for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Universiteitsweg 99, 3584 CG Utrecht, The Netherlands.