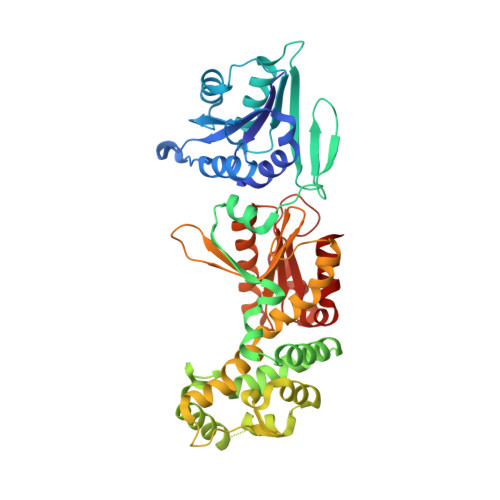
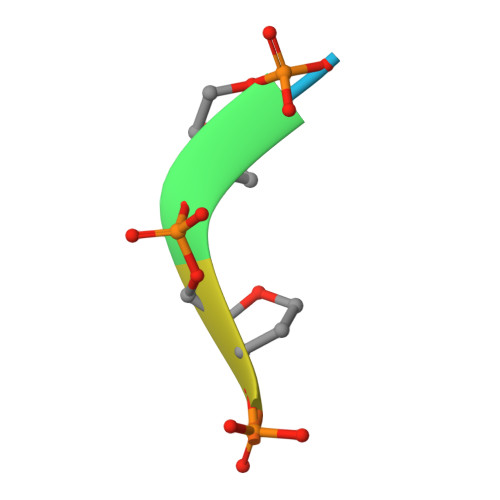
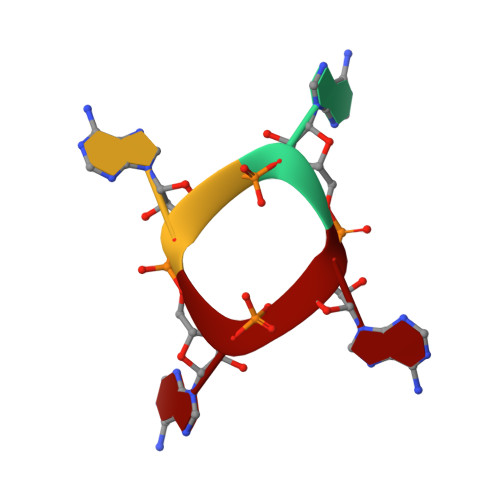
Substrate selectivity and catalytic activation of the type III CRISPR ancillary nuclease Can2.
Jungfer, K., Sigg, A., Jinek, M.(2024) Nucleic Acids Res 52: 462-473
- PubMed: 38033326
- DOI: https://doi.org/10.1093/nar/gkad1102
- Primary Citation of Related Structures:
8Q3Y, 8Q3Z, 8Q40, 8Q41, 8Q42, 8Q43, 8Q44 - PubMed Abstract:
Type III CRISPR-Cas systems provide adaptive immunity against foreign mobile genetic elements through RNA-guided interference. Sequence-specific recognition of RNA targets by the type III effector complex triggers the generation of cyclic oligoadenylate (cOA) second messengers that activate ancillary effector proteins, thus reinforcing the host immune response. The ancillary nuclease Can2 is activated by cyclic tetra-AMP (cA4); however, the mechanisms underlying cA4-mediated activation and substrate selectivity remain elusive. Here we report crystal structures of Thermoanaerobacter brockii Can2 (TbrCan2) in substrate- and product-bound complexes. We show that TbrCan2 is a single strand-selective DNase and RNase that binds substrates via a conserved SxTTS active site motif, and reveal molecular interactions underpinning its sequence preference for CA dinucleotides. Furthermore, we identify a molecular interaction relay linking the cA4 binding site and the nuclease catalytic site to enable divalent metal cation coordination and catalytic activation. These findings provide key insights into the molecular mechanisms of Can2 nucleases in type III CRISPR-Cas immunity and may guide their technological development for nucleic acid detection applications.
Organizational Affiliation:
Department of Biochemistry, University of Zurich, Zurich, Switzerland.