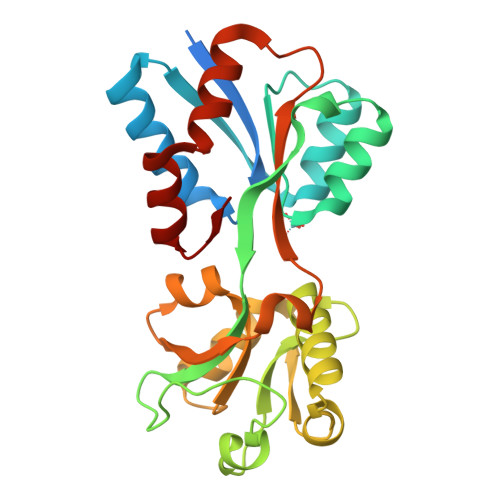
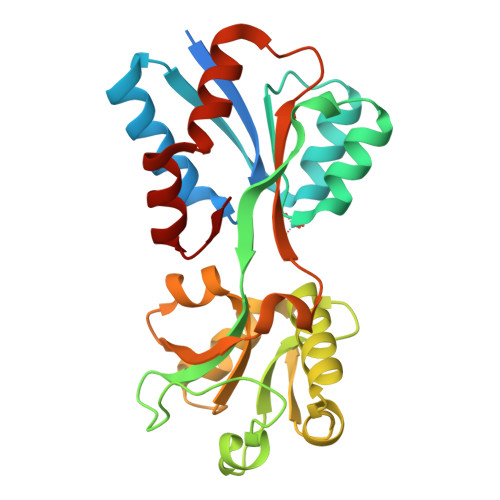
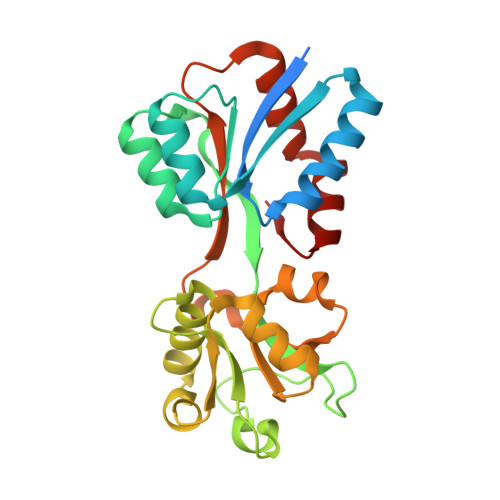
Structural analysis of molybdate binding protein ModA from Klebsiella pneumoniae.
Zhao, Q., Su, X., Wang, Y., Liu, R., Bartlam, M.(2023) Biochem Biophys Res Commun 681: 41-46
- PubMed: 37751633
- DOI: https://doi.org/10.1016/j.bbrc.2023.09.055
- Primary Citation of Related Structures:
8K8K, 8K8L - PubMed Abstract:
Klebsiella pneumoniae, a facultative anaerobe, relies on acquiring molybdenum to sustain growth in anaerobic conditions, a crucial factor for the pathogen to establish infections within host environments. Molybdenum plays a critical role in pathogenesis as it forms an essential component of cofactors for molybdoenzymes. K. pneumoniae utilizes the ABC (ATP-Binding-Cassette) transporter encoded by the modABC operon for uptake of the group VI elements molybdenum and tungsten. In this study, we determined the X-ray crystal structures of both the molybdenum-free and molybdenum-bound substrate-binding protein (SBP) ModA from Klebsiella pneumoniae to 2.00 Å and 1.77 Å resolution respectively. ModA crystallizes in the space group P222 with a single monomer in one asymmetric unit. The purified protein remained soluble and specifically bound molybdate and tungstate with K d values of 6.3 nM and 5.2 nM, respectively. Tungstate competes with molybdate by binding to ModA, resulting in enhanced antimicrobial activity. These data provide a starting point for structural and functional analyses of molybdate transport in K. pneumoniae.
Organizational Affiliation:
College of Life Sciences, Nankai University, Tianjin, 300071, China; State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, 300071, China.