Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Zhao, S., Olmayev-Yaakobov, D., Ru, W., Li, S., Chen, X., Zhang, J., Yao, X., Koren, I., Zhang, K., Xu, C.(2023) Proc Natl Acad Sci U S A 120: e2308870120-e2308870120
- PubMed: 37844242
- DOI: https://doi.org/10.1073/pnas.2308870120
- Primary Citation of Related Structures:
8JAL, 8JAQ, 8JAR, 8JAS, 8JAU, 8JAV - PubMed Abstract:
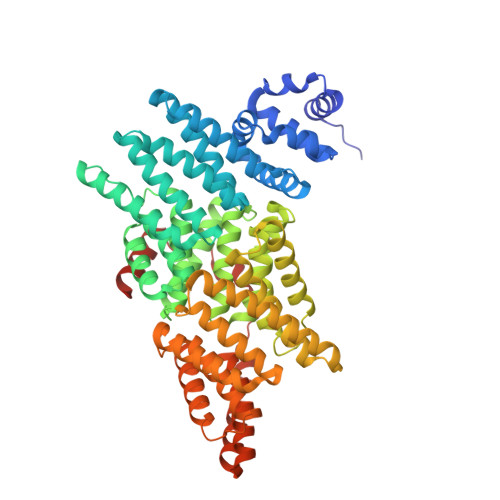
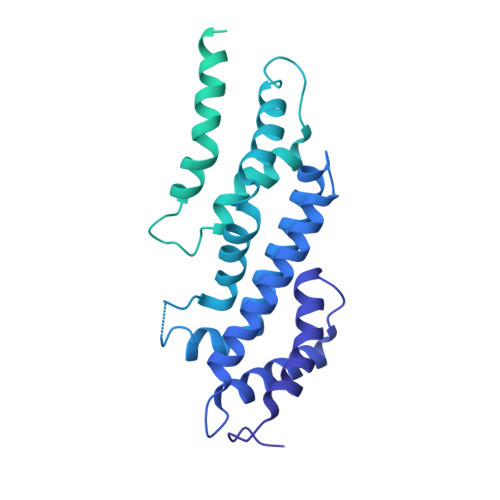
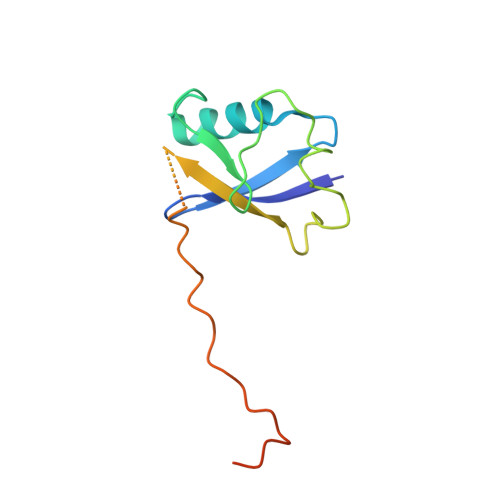
E3 ubiquitin ligases determine the specificity of eukaryotic protein degradation by selective binding to destabilizing protein motifs, termed degrons, in substrates for ubiquitin-mediated proteolysis. The exposed C-terminal residues of proteins can act as C-degrons that are recognized by distinct substrate receptors (SRs) as part of dedicated cullin-RING E3 ubiquitin ligase (CRL) complexes. APPBP2, an SR of Cullin 2-RING ligase (CRL2), has been shown to recognize R-x-x-G/C-degron; however, the molecular mechanism of recognition remains elusive. By solving several cryogenic electron microscopy structures of active CRL2 APPBP2 bound with different R-x-x-G/C-degrons, we unveiled the molecular mechanisms underlying the assembly of the CRL2 APPBP2 dimer and tetramer, as well as C-degron recognition. The structural study, complemented by binding experiments and cell-based assays, demonstrates that APPBP2 specifically recognizes the R-x-x-G/C-degron via a bipartite mechanism; arginine and glycine, which play critical roles in C-degron recognition, accommodate distinct pockets that are spaced by two residues. In addition, the binding pocket is deep enough to enable the interaction of APPBP2 with the motif placed at or up to three residues upstream of the C-end. Overall, our study not only provides structural insight into CRL2 APPBP2 -mediated protein turnover but also serves as the basis for future structure-based chemical probe design.
Organizational Affiliation:
Ministry of Education Key Laboratory for Cellular Dynamics, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027, Peoples Republic of China.