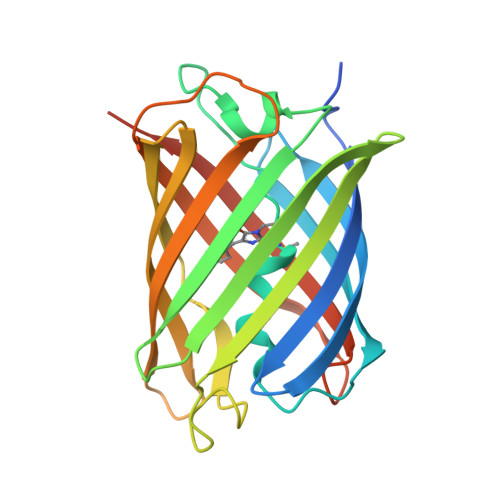
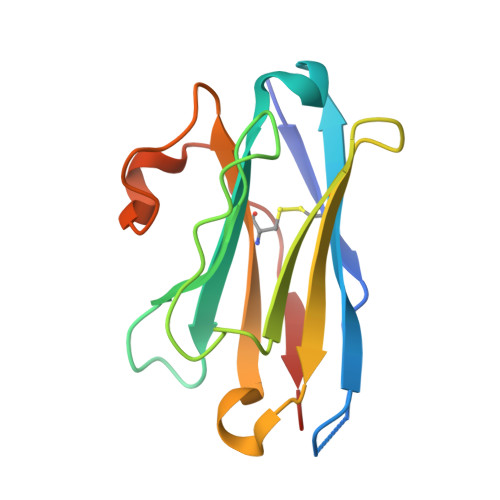
Structural Insights into the Binding of Red Fluorescent Protein mCherry-Specific Nanobodies.
Liang, H., Ma, Z., Wang, Z., Zhong, P., Li, R., Jiang, H., Zong, X., Zhong, C., Liu, X., Liu, P., Liu, J., Zhu, H., Liu, R., Ding, Y.(2023) Int J Mol Sci 24
- PubMed: 37108116
- DOI: https://doi.org/10.3390/ijms24086952
- Primary Citation of Related Structures:
8ILX, 8IM0, 8IM1 - PubMed Abstract:
Red fluorescent proteins (RFPs) have broad applications in life science research, and the manipulation of RFPs using nanobodies can expand their potential uses. However, the structural information available for nanobodies that bind with RFPs is still insufficient. In this study, we cloned, expressed, purified, and crystallized complexes formed by mCherry with LaM1, LaM3, and LaM8. Then, we analyzed the biochemical properties of the complexes using mass spectrometry (MS), fluorescence-detected size exclusion chromatography (FSEC), isothermal titration calorimetry (ITC), and bio-layer interferometry (BLI) technology. We determined the crystal structure of mCherry-LaM1, mCherry-LaM3, and mCherry-LaM8, with resolutions of 2.05 Å, 3.29 Å, and 1.31 Å, respectively. In this study, we systematically compared various parameters of several LaM series nanobodies, including LaM1, LaM3, and LaM8, with previously reported data on LaM2, LaM4, and LaM6, specifically examining their structural information. After designing multivalent tandem LaM1-LaM8 and LaM8-LaM4 nanobodies based on structural information, we characterized their properties, revealing their higher affinity and specificity to mCherry. Our research provides novel structural insights that could aid in understanding nanobodies targeting a specific target protein. This could provide a starting point for developing enhanced mCherry manipulation tools.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200438, China.