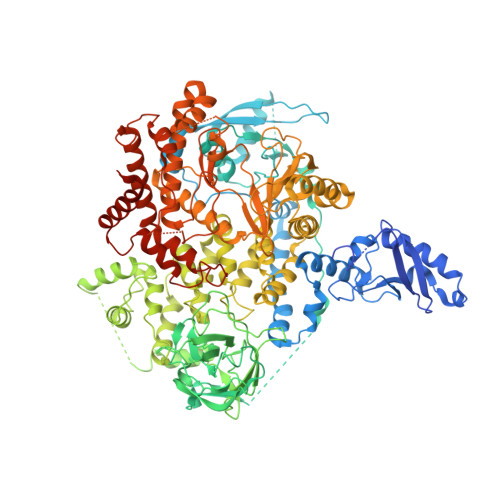
Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Zhou, Q., Liu, X., Neri, D., Li, W., Favalli, N., Bassi, G., Yang, S., Yang, D., Vogt, P.K., Wang, M.W.(2023) Proc Natl Acad Sci U S A 120: e2304071120-e2304071120
- PubMed: 37585458
- DOI: https://doi.org/10.1073/pnas.2304071120
- Primary Citation of Related Structures:
8ILR, 8ILS, 8ILV - PubMed Abstract:
Class IA phosphoinositide 3-kinase alpha (PI3Kα) is an important drug target because it is one of the most frequently mutated proteins in human cancers. However, small molecule inhibitors currently on the market or under development have safety concerns due to a lack of selectivity. Therefore, other chemical scaffolds or unique mechanisms of catalytic kinase inhibition are needed. Here, we report the cryo-electron microscopy structures of wild-type PI3Kα, the dimer of p110α and p85α, in complex with three Y-shaped ligands [cpd16 (compound 16), cpd17 (compound 17), and cpd18 (compound 18)] of different affinities and no inhibitory effect on the kinase activity. Unlike ATP-competitive inhibitors, cpd17 adopts a Y-shaped conformation with one arm inserted into a binding pocket formed by R770 and W780 and the other arm lodged in the ATP-binding pocket at an angle that is different from that of the ATP phosphate tail. Such a special interaction induces a conformation of PI3Kα resembling that of the unliganded protein. These observations were confirmed with two isomers (cpd16 and cpd18). Further analysis of these Y-shaped ligands revealed the structural basis of differential binding affinities caused by stereo- or regiochemical modifications. Our results may offer a different direction toward the design of therapeutic agents against PI3Kα.
Organizational Affiliation:
Department of Pharmacology, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.