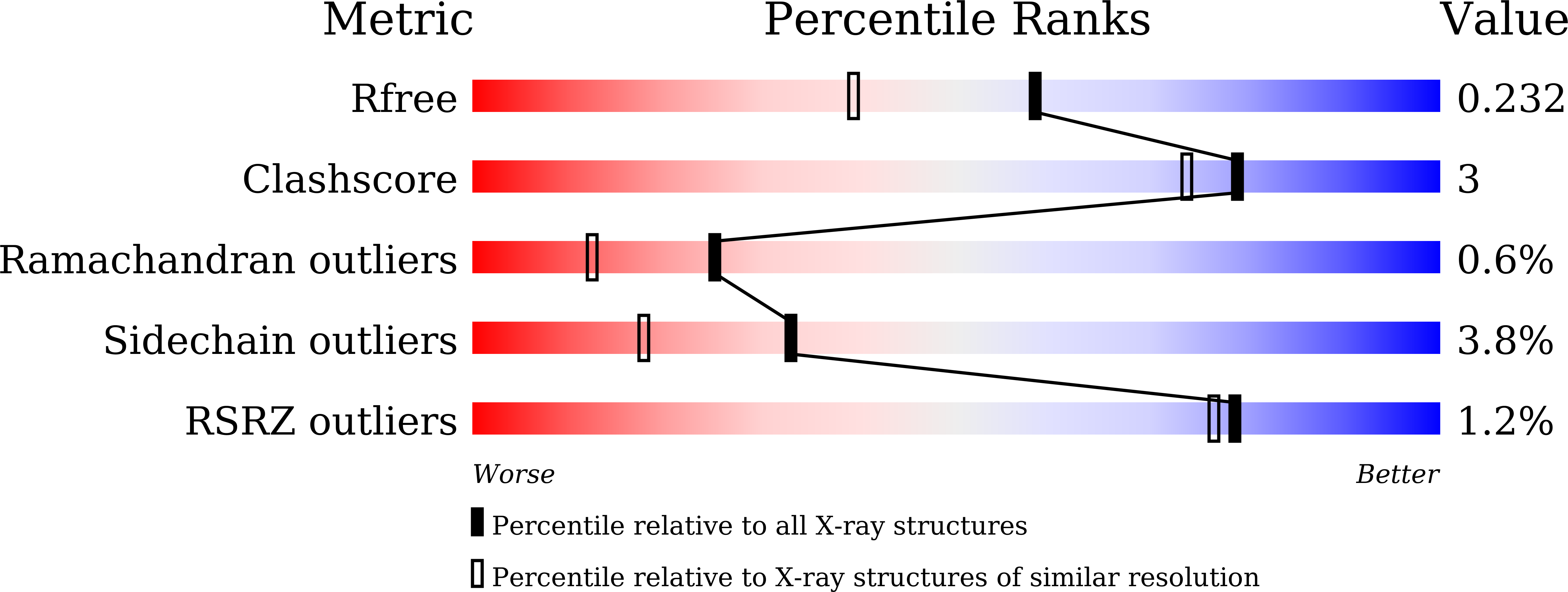
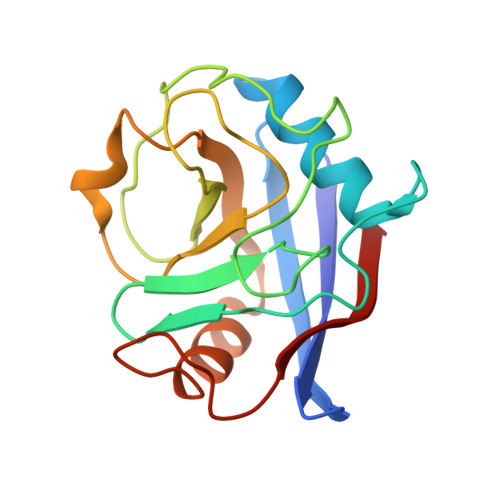
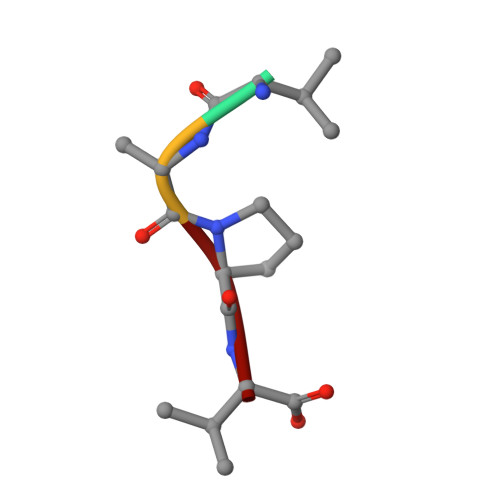
PPIA dictates NRF2 stability to promote lung cancer progression.
Lu, W., Cui, J., Wang, W., Hu, Q., Xue, Y., Liu, X., Gong, T., Lu, Y., Ma, H., Yang, X., Feng, B., Wang, Q., Zhang, N., Xu, Y., Liu, M., Nussinov, R., Cheng, F., Ji, H., Huang, J.(2024) Nat Commun 15: 4703-4703
- PubMed: 38830868
- DOI: https://doi.org/10.1038/s41467-024-48364-4
- Primary Citation of Related Structures:
8HZ8 - PubMed Abstract:
Nuclear factor erythroid 2-related factor 2 (NRF2) hyperactivation has been established as an oncogenic driver in a variety of human cancers, including non-small cell lung cancer (NSCLC). However, despite massive efforts, no specific therapy is currently available to target NRF2 hyperactivation. Here, we identify peptidylprolyl isomerase A (PPIA) is required for NRF2 protein stability. Ablation of PPIA promotes NRF2 protein degradation and blocks NRF2-driven growth in NSCLC cells. Mechanistically, PPIA physically binds to NRF2 and blocks the access of ubiquitin/Kelch Like ECH Associated Protein 1 (KEAP1) to NRF2, thus preventing ubiquitin-mediated degradation. Our X-ray co-crystal structure reveals that PPIA directly interacts with a NRF2 interdomain linker via a trans-proline 174-harboring hydrophobic sequence. We further demonstrate that an FDA-approved drug, cyclosporin A (CsA), impairs the interaction of NRF2 with PPIA, inducing NRF2 ubiquitination and degradation. Interestingly, CsA interrupts glutamine metabolism mediated by the NRF2/KLF5/SLC1A5 pathway, consequently suppressing the growth of NRF2-hyperactivated NSCLC cells. CsA and a glutaminase inhibitor combination therapy significantly retard tumor progression in NSCLC patient-derived xenograft (PDX) models with NRF2 hyperactivation. Our study demonstrates that targeting NRF2 protein stability is an actionable therapeutic approach to treat NRF2-hyperactivated NSCLC.
Organizational Affiliation:
Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai, China. wqlu@bio.ecnu.edu.cn.