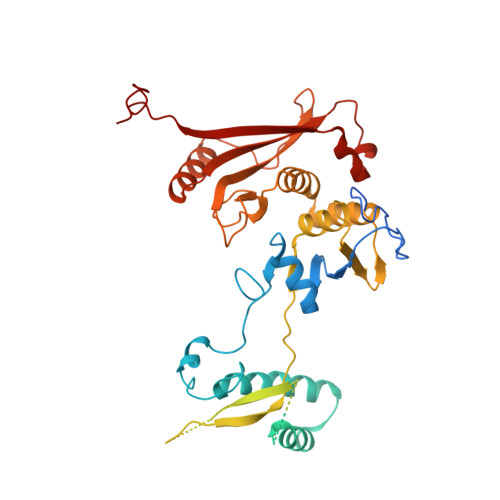
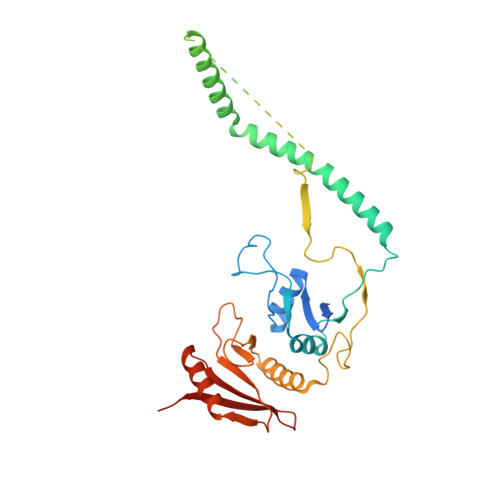
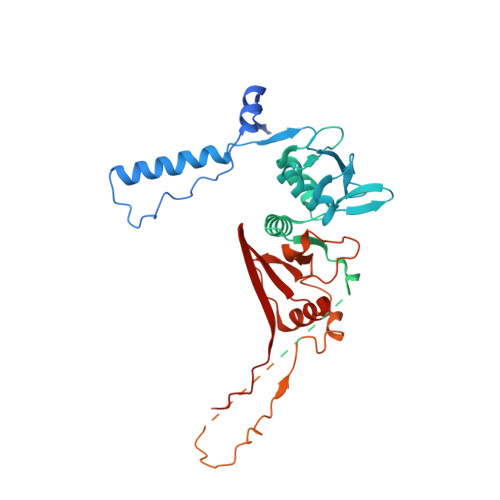
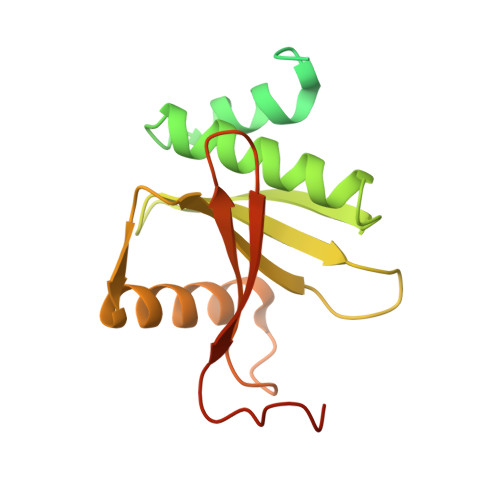
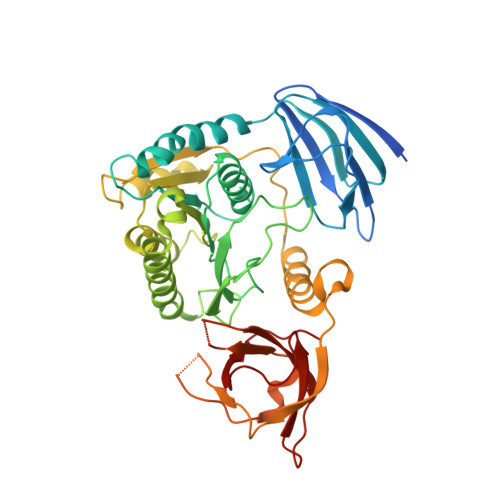
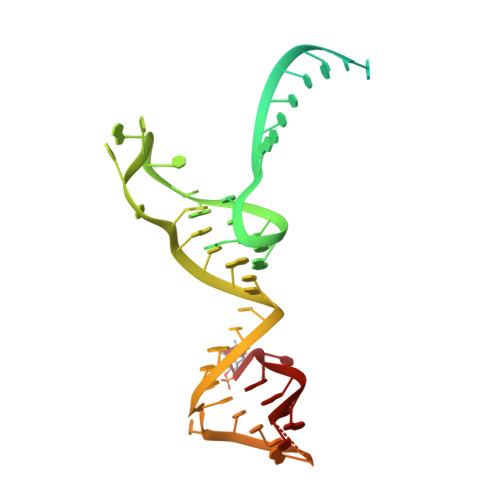
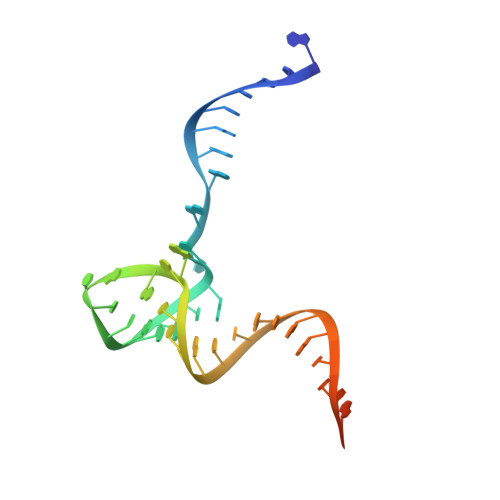
Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Zhang, X., Yang, F., Zhan, X., Bian, T., Xing, Z., Lu, Y., Shi, Y.(2023) Mol Cell 83: 1328-1339.e4
- PubMed: 37028420
- DOI: https://doi.org/10.1016/j.molcel.2023.03.015
- Primary Citation of Related Structures:
8HMY, 8HMZ - PubMed Abstract:
Removal of the intron from precursor-tRNA (pre-tRNA) is essential in all three kingdoms of life. In humans, this process is mediated by the tRNA splicing endonuclease (TSEN) comprising four subunits: TSEN2, TSEN15, TSEN34, and TSEN54. Here, we report the cryo-EM structures of human TSEN bound to full-length pre-tRNA in the pre-catalytic and post-catalytic states at average resolutions of 2.94 and 2.88 Å, respectively. Human TSEN features an extended surface groove that holds the L-shaped pre-tRNA. The mature domain of pre-tRNA is recognized by conserved structural elements of TSEN34, TSEN54, and TSEN2. Such recognition orients the anticodon stem of pre-tRNA and places the 3'-splice site and 5'-splice site into the catalytic centers of TSEN34 and TSEN2, respectively. The bulk of the intron sequences makes no direct interaction with TSEN, explaining why pre-tRNAs of varying introns can be accommodated and cleaved. Our structures reveal the molecular ruler mechanism of pre-tRNA cleavage by TSEN.
Organizational Affiliation:
Research Center for Industries of the Future, Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Institute of Biology, Westlake Institute for Advanced Study, 18 Shilongshan Road, Hangzhou 310024, Zhejiang, China; Westlake Laboratory of Life Sciences and Biomedicine, 18 Shilongshan Road, Hangzhou 310024, Zhejiang, China. Electronic address: xiaofengzhang@ustc.edu.cn.