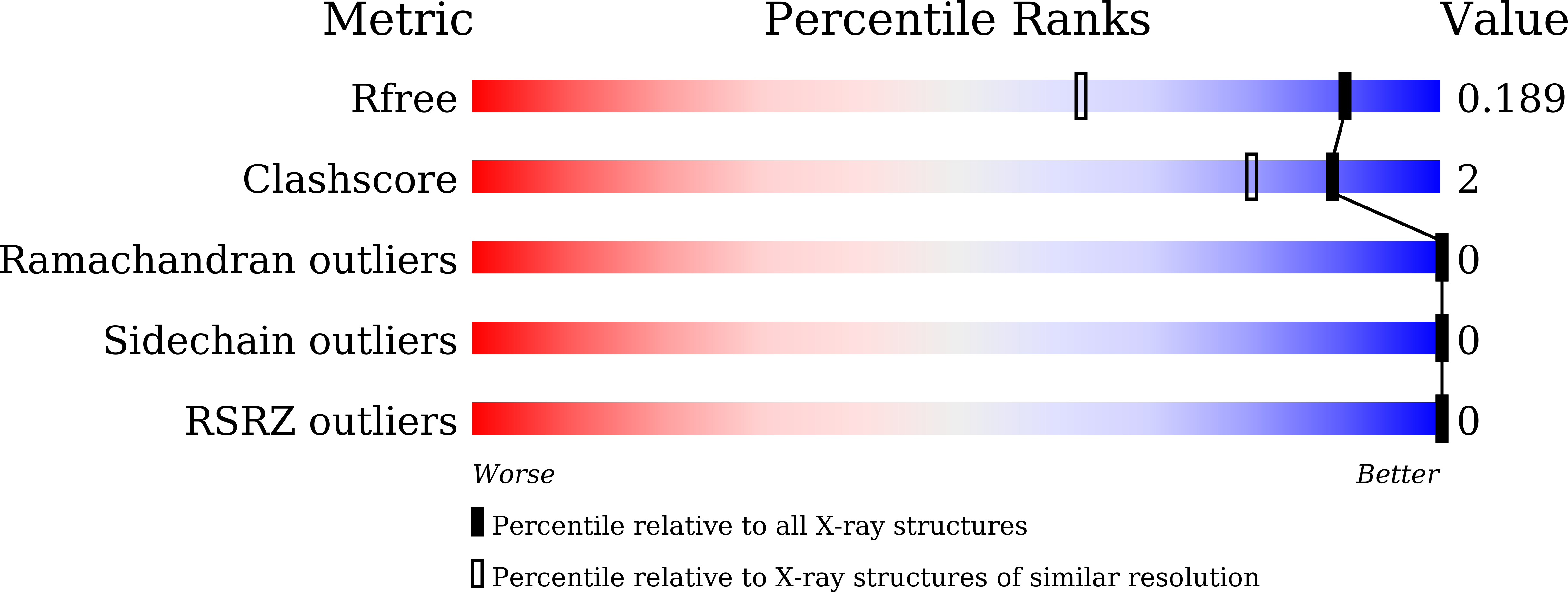
Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Kim, J.H., Pandit, N., Yoo, M., Park, T.H., Choi, J.U., Park, C.H., Jung, K.Y., Lee, B.I.(2023) Sci Rep 13: 10805-10805
- PubMed: 37402749
- DOI: https://doi.org/10.1038/s41598-023-37527-w
- Primary Citation of Related Structures:
7YMG, 7YQ9, 8GPZ, 8GQ0 - PubMed Abstract:
BRD4 contains two tandem bromodomains (BD1 and BD2) that recognize acetylated lysine for epigenetic reading, and these bromodomains are promising therapeutic targets for treating various diseases, including cancers. BRD4 is a well-studied target, and many chemical scaffolds for inhibitors have been developed. Research on the development of BRD4 inhibitors against various diseases is actively being conducted. Herein, we propose a series of [1,2,4]triazolo[4,3-b]pyridazine derivatives as bromodomain inhibitors with micromolar IC 50 values. We characterized the binding modes by determining the crystal structures of BD1 in complex with four selected inhibitors. Compounds containing [1,2,4] triazolo[4,3-b]pyridazine derivatives offer promising starting molecules for designing potent BRD4 BD inhibitors.
Organizational Affiliation:
Research Institute, National Cancer Center, Goyang, Gyeonggi, 10408, Republic of Korea.