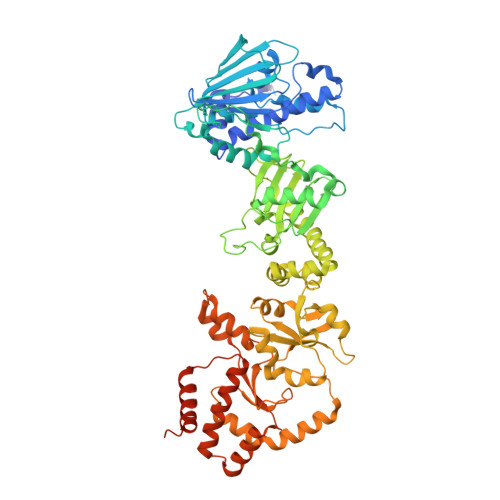
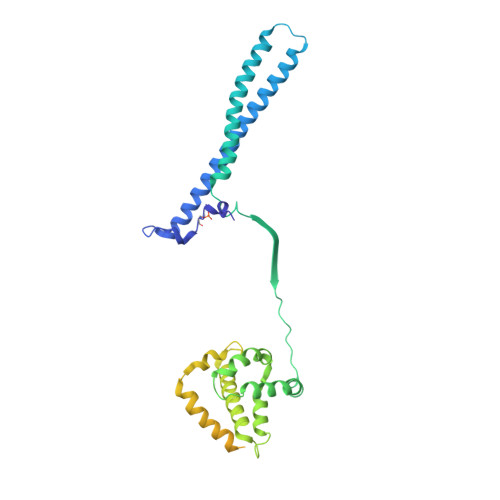
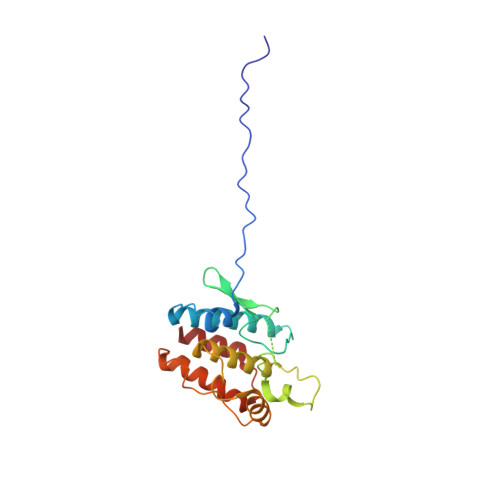
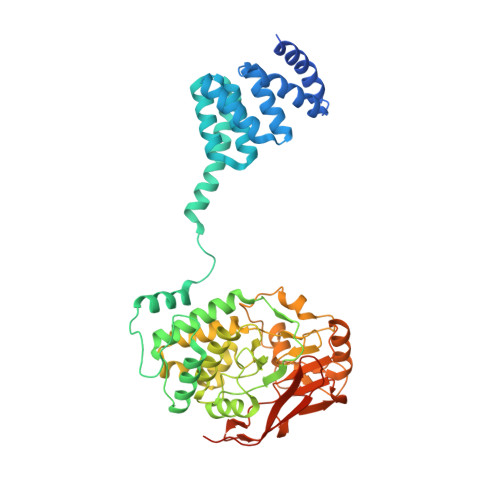
Hsp90 provides a platform for kinase dephosphorylation by PP5.
Jaime-Garza, M., Nowotny, C.A., Coutandin, D., Wang, F., Tabios, M., Agard, D.A.(2023) Nat Commun 14: 2197-2197
- PubMed: 37069154
- DOI: https://doi.org/10.1038/s41467-023-37659-7
- Primary Citation of Related Structures:
8GAE, 8GFT - PubMed Abstract:
The Hsp90 molecular chaperone collaborates with the phosphorylated Cdc37 cochaperone for the folding and activation of its many client kinases. As with many kinases, the Hsp90 client kinase CRaf is activated by phosphorylation at specific regulatory sites. The cochaperone phosphatase PP5 dephosphorylates CRaf and Cdc37 in an Hsp90-dependent manner. Although dephosphorylating Cdc37 has been proposed as a mechanism for releasing Hsp90-bound kinases, here we show that Hsp90 bound kinases sterically inhibit Cdc37 dephosphorylation indicating kinase release must occur before Cdc37 dephosphorylation. Our cryo-EM structure of PP5 in complex with Hsp90:Cdc37:CRaf reveals how Hsp90 both activates PP5 and scaffolds its association with the bound CRaf to dephosphorylate phosphorylation sites neighboring the kinase domain. Thus, we directly show how Hsp90's role in maintaining protein homeostasis goes beyond folding and activation to include post translationally modifying its client kinases.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of California, San Francisco, San Francisco, CA, 94143, USA.