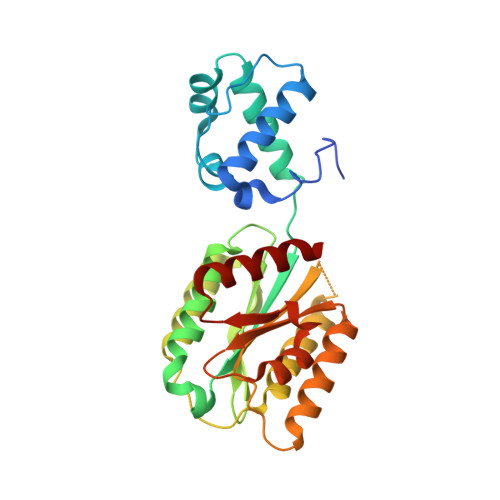
Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Gajjar, P.L., Pedroza Romo, M.J., Litchfield, C.M., Callahan, M., Redd, N., Nawarathnage, S., Soleimani, S., Averett, J., Wilson, E., Lewis, A., Stewart, C., Tseng, Y.J., Doukov, T., Lebedev, A., Moody, J.D.(2023) Acta Crystallogr D Struct Biol 79: 925-943
- PubMed: 37747038
- DOI: https://doi.org/10.1107/S2059798323007246
- Primary Citation of Related Structures:
8FT6, 8FT8, 8FZ4, 8FZU, 8FZV - PubMed Abstract:
TELSAM-fusion crystallization has the potential to become a revolutionary tool for the facile crystallization of proteins. TELSAM fusion can increase the crystallization rate and enable crystallization at low protein concentrations, in some cases with minimal crystal contacts [Nawarathnage et al. (2022), Open Biol. 12, 210271]. Here, requirements for the linker composition between 1TEL and a fused CMG2 vWa domain were investigated. Ala-Ala, Ala-Val, Thr-Val and Thr-Thr linkers were evaluated, comparing metrics for crystallization propensity and crystal order. The effect on crystallization of removing or retaining the purification tag was then tested. It was discovered that increasing the linker bulk and retaining the 10×His purification tag improved the diffraction resolution, likely by decreasing the number of possible vWa-domain orientations in the crystal. Additionally, it was discovered that some vWa-domain binding modes are correlated with scrambling of the 1TEL polymer orientation in crystals and an effective mitigation strategy for this pathology is presented.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Brigham Young University, 701 East University Parkway, Provo, UT 84602, USA.