Insights into the mechanism of SARS-CoV-2 main protease autocatalytic maturation from model precursors.
Aniana, A., Nashed, N.T., Ghirlando, R., Coates, L., Kneller, D.W., Kovalevsky, A., Louis, J.M.(2023) Commun Biol 6: 1159-1159
- PubMed: 37957287
- DOI: https://doi.org/10.1038/s42003-023-05469-8
- Primary Citation of Related Structures:
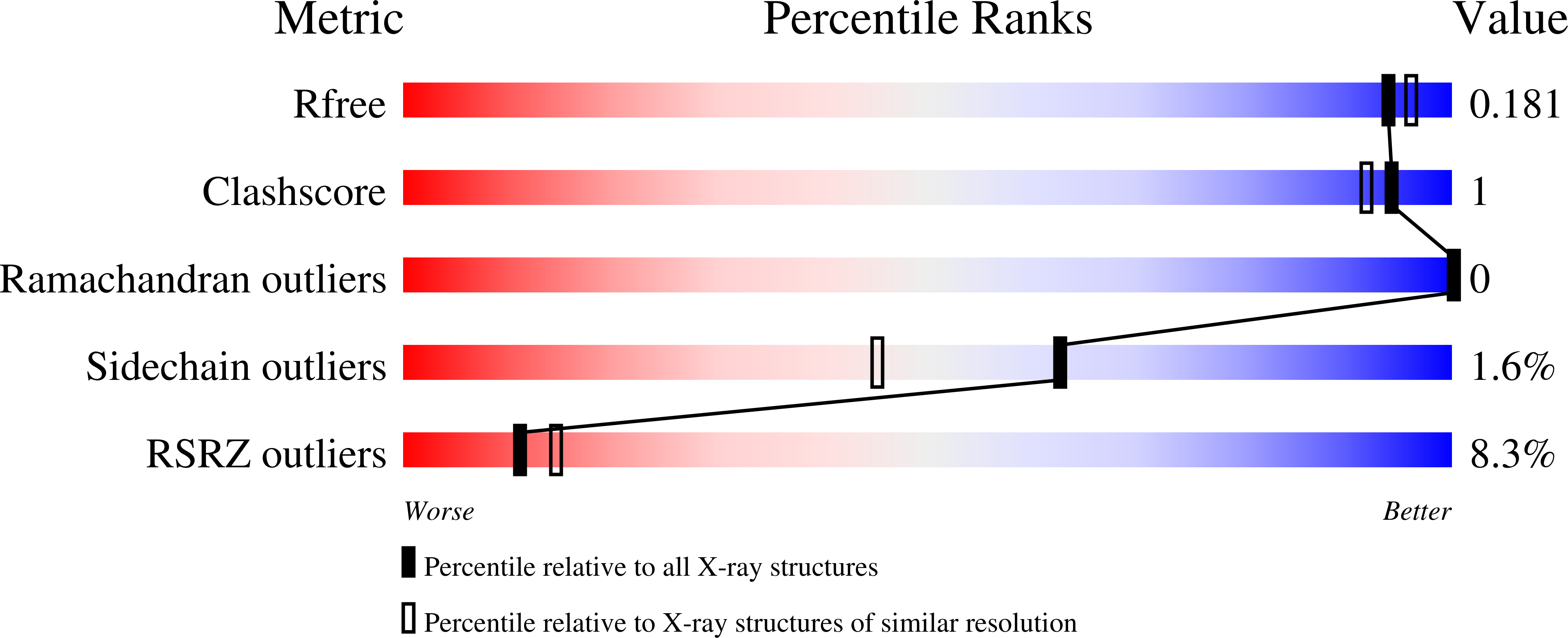
8FIG - PubMed Abstract:
A critical step for SARS-CoV-2 assembly and maturation involves the autoactivation of the main protease (MPro WT ) from precursor polyproteins. Upon expression, a model precursor of MPro WT mediates its own release at its termini rapidly to yield a mature dimer. A construct with an E290A mutation within MPro exhibits time dependent autoprocessing of the accumulated precursor at the N-terminal nsp4/nsp5 site followed by the C-terminal nsp5/nsp6 cleavage. In contrast, a precursor containing E290A and R298A mutations (MPro M ) displays cleavage only at the nsp4/nsp5 site to yield an intermediate monomeric product, which is cleaved at the nsp5/nsp6 site only by MPro WT . MPro M and the catalytic domain (MPro 1-199 ) fused to the truncated nsp4 region also show time-dependent conversion in vitro to produce MPro M and MPro 1-199 , respectively. The reactions follow first-order kinetics indicating that the nsp4/nsp5 cleavage occurs via an intramolecular mechanism. These results support a mechanism involving an N-terminal intramolecular cleavage leading to an increase in the dimer population and followed by an intermolecular cleavage at the C-terminus. Thus, targeting the predominantly monomeric MPro precursor for inhibition may lead to the identification of potent drugs for treatment.
Organizational Affiliation:
Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, DHHS, Bethesda, MD, 20892-0520, USA.