Molecular Bidents with Two Electrophilic Warheads as a New Pharmacological Modality.
Li, Z., Jiang, J., Ficarro, S.B., Beyett, T.S., To, C., Tavares, I., Zhu, Y., Li, J., Eck, M.J., Janne, P.A., Marto, J.A., Zhang, T., Che, J., Gray, N.S.(2024) ACS Cent Sci 10: 1156-1166
- PubMed: 38947214
- DOI: https://doi.org/10.1021/acscentsci.3c01245
- Primary Citation of Related Structures:
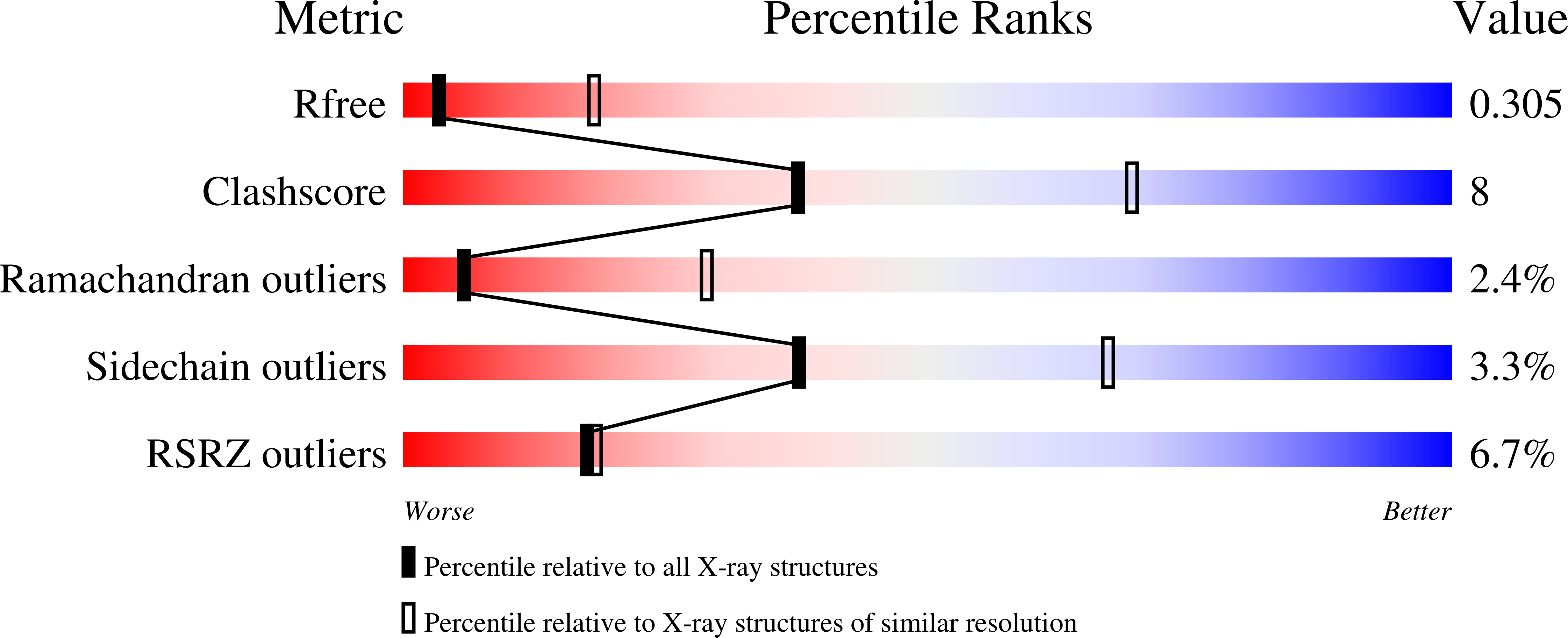
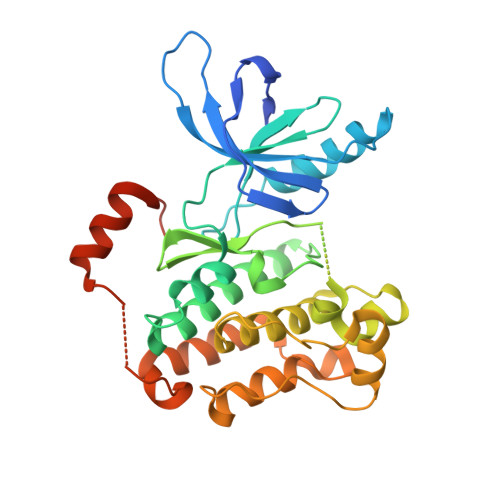
8EME - PubMed Abstract:
A systematic strategy to develop dual-warhead inhibitors is introduced to circumvent the limitations of conventional covalent inhibitors such as vulnerability to mutations of the corresponding nucleophilic residue. Currently, all FDA-approved covalent small molecules feature one electrophile, leaving open a facile route to acquired resistance. We conducted a systematic analysis of human proteins in the protein data bank to reveal ∼400 unique targets amendable to dual covalent inhibitors, which we term "molecular bidents". We demonstrated this strategy by targeting two kinases: MKK7 and EGFR. The designed compounds, ZNL-8162 and ZNL-0056, are ATP-competitive inhibitors that form two covalent bonds with cysteines and retain potency against single cysteine mutants. Therefore, molecular bidents represent a new pharmacological modality with the potential for improved selectivity, potency, and drug resistance profile.
Organizational Affiliation:
Department of Chemical and Systems Biology, Stanford Cancer Institute, ChEM-H, Stanford University, Stanford, California 94305, United States.