Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
Donovan, K.A., Coombes, D., Dobson, R.C.J., Cooper, T.F.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Pyruvate kinase | 470 | Escherichia coli | Mutation(s): 1 Gene Names: pykF EC: 2.7.1.40 | 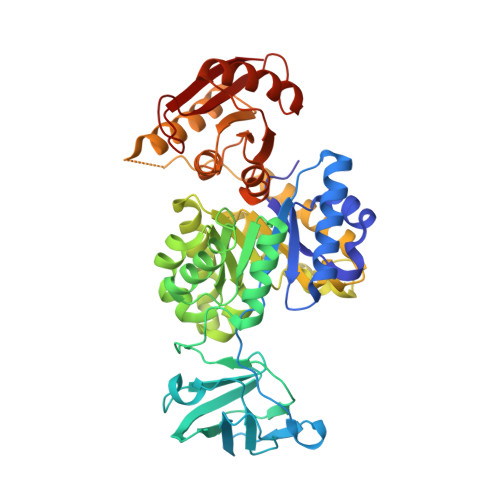 | |
UniProt | |||||
Find proteins for P0AD61 (Escherichia coli (strain K12)) Explore P0AD61 Go to UniProtKB: P0AD61 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AD61 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 74.68 | α = 90 |
| b = 247.412 | β = 90 |
| c = 129.965 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | US Army Research Office W911NF-11-1-0481 | |
| National Science Foundation (NSF, United States) | United States | DEB-1253650 |