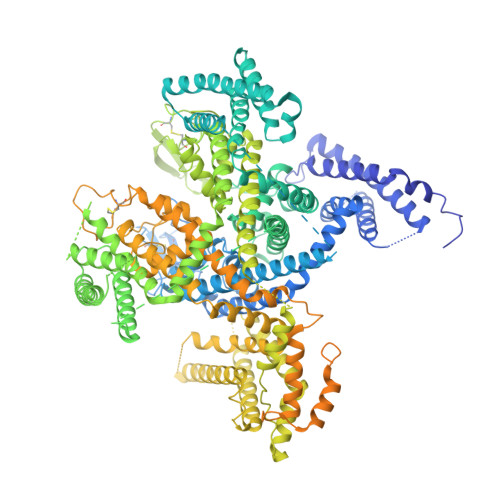
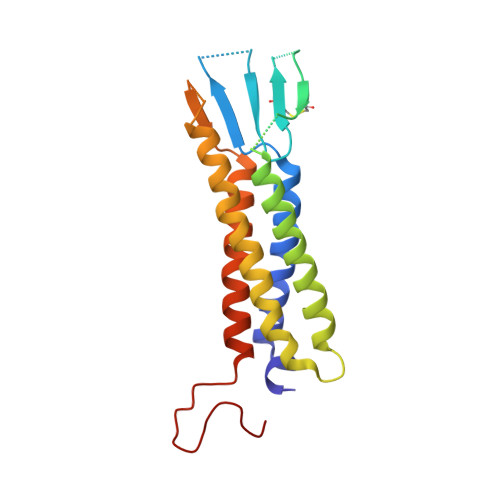
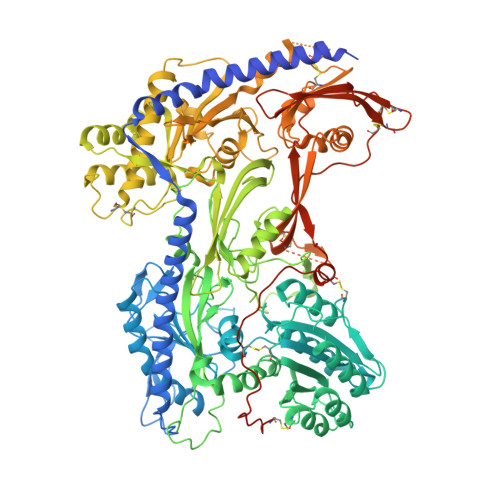
Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Yao, X., Gao, S., Wang, J., Li, Z., Huang, J., Wang, Y., Wang, Z., Chen, J., Fan, X., Wang, W., Jin, X., Pan, X., Yu, Y., Lagrutta, A., Yan, N.(2022) Cell 185: 4801-4810.e13
- PubMed: 36417914
- DOI: https://doi.org/10.1016/j.cell.2022.10.024
- Primary Citation of Related Structures:
8E56, 8E57, 8E58, 8E59, 8E5A, 8E5B - PubMed Abstract:
Drug-drug interaction of the antiviral sofosbuvir and the antiarrhythmics amiodarone has been reported to cause fatal heartbeat slowing. Sofosbuvir and its analog, MNI-1, were reported to potentiate the inhibition of cardiomyocyte calcium handling by amiodarone, which functions as a multi-channel antagonist, and implicate its inhibitory effect on L-type Ca v channels, but the molecular mechanism has remained unclear. Here we present systematic cryo-EM structural analysis of Ca v 1.1 and Ca v 1.3 treated with amiodarone or sofosbuvir alone, or sofosbuvir/MNI-1 combined with amiodarone. Whereas amiodarone alone occupies the dihydropyridine binding site, sofosbuvir is not found in the channel when applied on its own. In the presence of amiodarone, sofosbuvir/MNI-1 is anchored in the central cavity of the pore domain through specific interaction with amiodarone and directly obstructs the ion permeation path. Our study reveals the molecular basis for the physical, pharmacodynamic interaction of two drugs on the scaffold of Ca v channels.
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Princeton, NJ 08544, USA.