Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Thakur, A.K., Miller, S.E., Liau, N.P.D., Hwang, S., Hansen, S., de Sousa E Melo, F., Sudhamsu, J., Hannoush, R.N.(2023) ACS Chem Biol 18: 772-784
- PubMed: 36893429
- DOI: https://doi.org/10.1021/acschembio.2c00753
- Primary Citation of Related Structures:
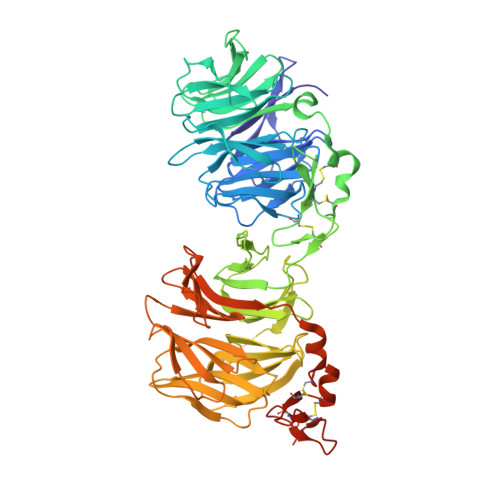
8DVL, 8DVM, 8DVN - PubMed Abstract:
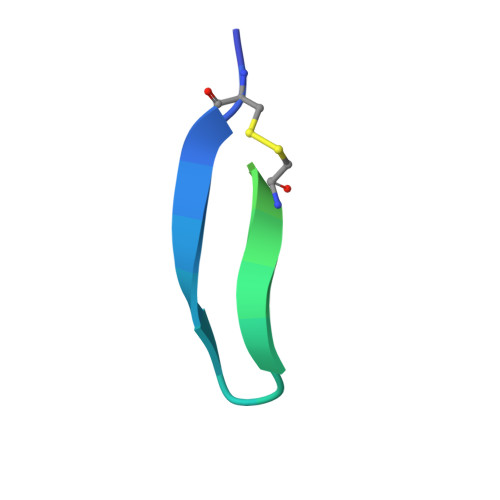
Wnt ligands are critical for tissue homeostasis and form a complex with LRP6 and frizzled coreceptors to initiate Wnt/β-catenin signaling. Yet, how different Wnts achieve various levels of signaling activation through distinct domains on LRP6 remains elusive. Developing tool ligands that target individual LRP6 domains could help elucidate the mechanism of Wnt signaling regulation and uncover pharmacological approaches for pathway modulation. We employed directed evolution of a disulfide constrained peptide (DCP) to identify molecules that bind to the third β-propeller domain of LRP6. The DCPs antagonize Wnt3a while sparing Wnt1 signaling. Using PEG linkers with different geometries, we converted the Wnt3a antagonist DCPs to multivalent molecules that potentiated Wnt1 signaling by clustering the LRP6 coreceptor. The mechanism of potentiation is unique as it occurred only in the presence of extracellular secreted Wnt1 ligand. While all DCPs recognized a similar binding interface on LRP6, they displayed different spatial orientations that influenced their cellular activities. Moreover, structural analyses revealed that the DCPs exhibited new folds that were distinct from the parent DCP framework they were evolved from. The multivalent ligand design principles highlighted in this study provide a path for developing peptide agonists that modulate different branches of cellular Wnt signaling.
Organizational Affiliation:
Department of Early Discovery Biochemistry, Genentech, South San Francisco, California 94080, United States.