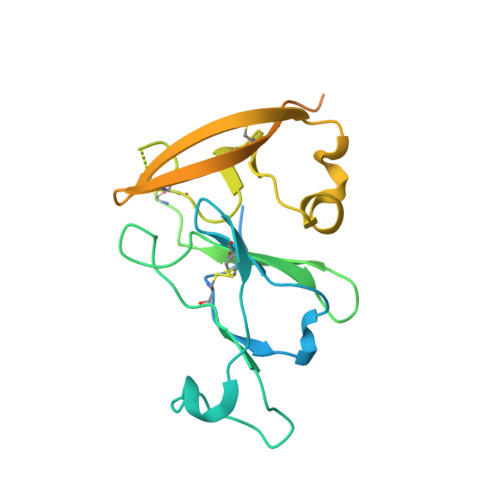
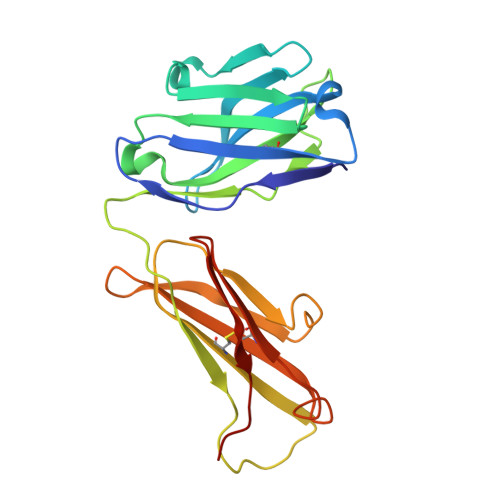
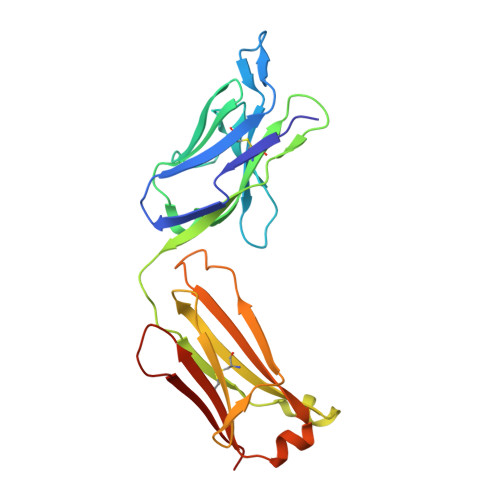
Regions of hepatitis C virus E2 required for membrane association.
Kumar, A., Rohe, T.C., Elrod, E.J., Khan, A.G., Dearborn, A.D., Kissinger, R., Grakoui, A., Marcotrigiano, J.(2023) Nat Commun 14: 433-433
- PubMed: 36702826
- DOI: https://doi.org/10.1038/s41467-023-36183-y
- Primary Citation of Related Structures:
8DK6 - PubMed Abstract:
Hepatitis C virus (HCV) uses a hybrid entry mechanism. Current structural data suggest that upon exposure to low pH and Cluster of Differentiation 81 (CD81), the amino terminus of envelope glycoprotein E2 becomes ordered and releases an internal loop with two invariant aromatic residues into the host membrane. Here, we present the structure of an amino-terminally truncated E2 with the membrane binding loop in a bent conformation and the aromatic side chains sequestered. Comparison with three previously reported E2 structures with the same Fab indicates that this internal loop is flexible, and that local context influences the exposure of hydrophobic residues. Biochemical assays show that the amino-terminally truncated E2 lacks the baseline membrane-binding capacity of the E2 ectodomain. Thus, the amino terminal region is a critical determinant for both CD81 and membrane interaction. These results provide new insights into the HCV entry mechanism.
Organizational Affiliation:
Structural Virology Section, Laboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, 20892, USA.