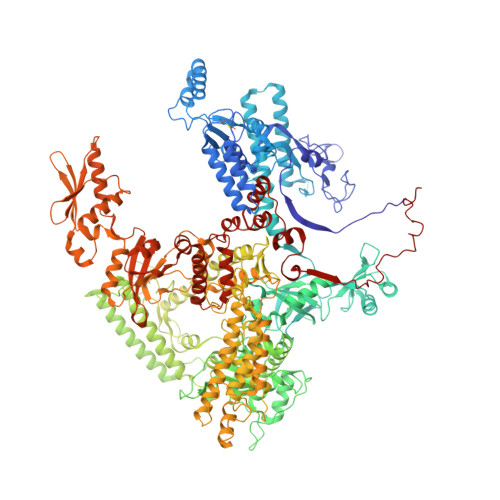
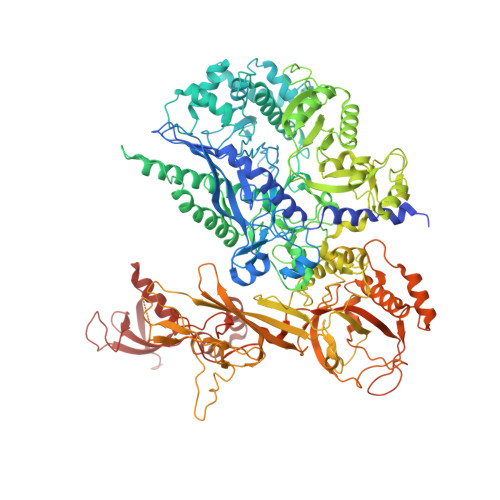
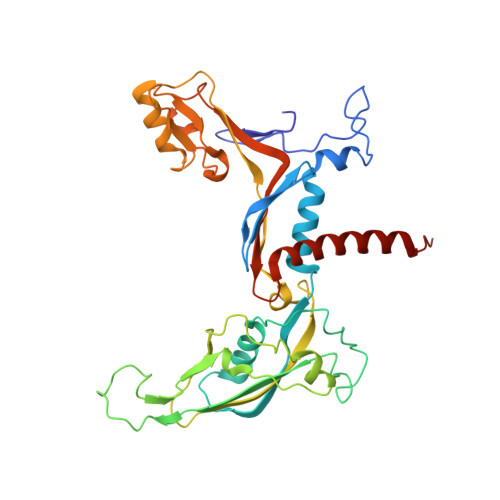
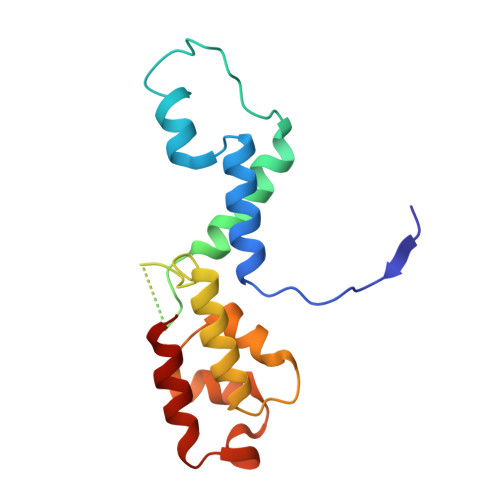
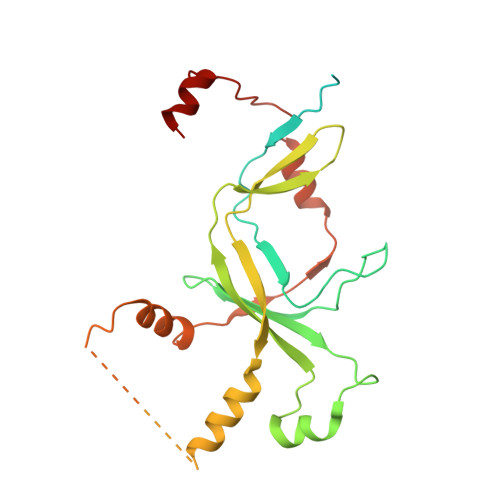
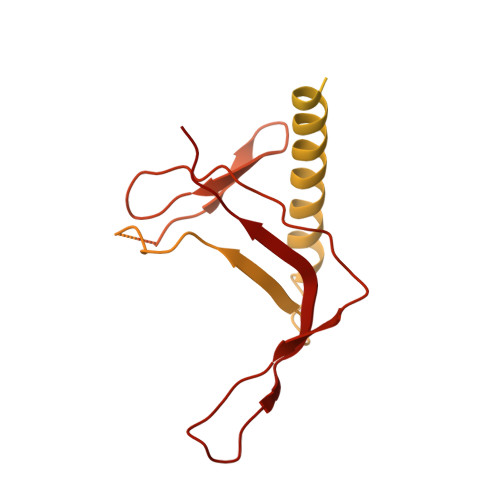
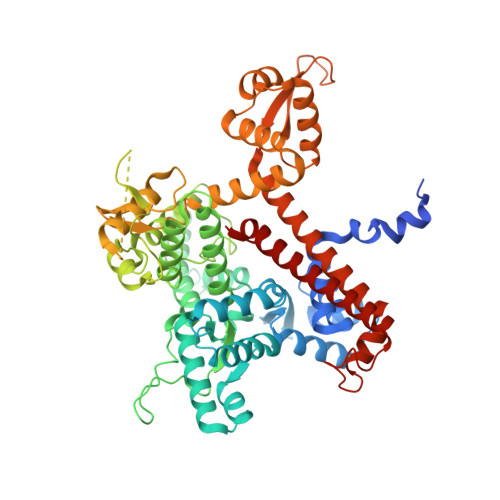
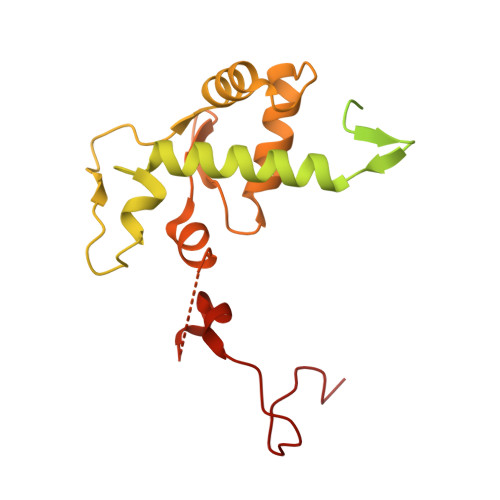
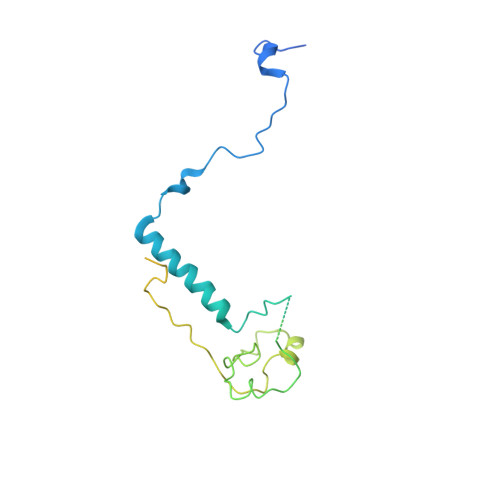
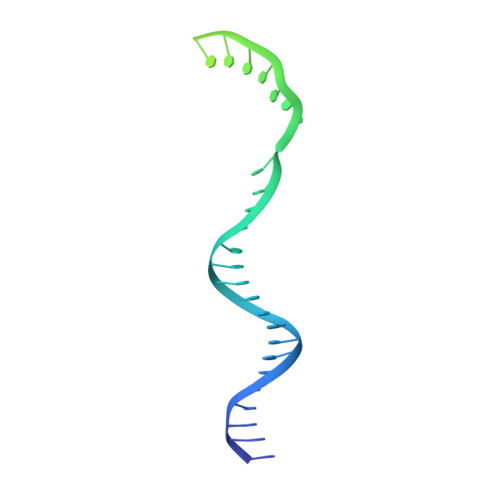
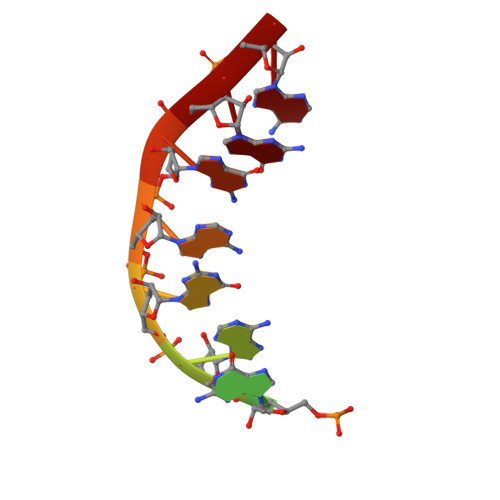
Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nguyen, P.Q., Huecas, S., Asif-Laidin, A., Plaza-Pegueroles, A., Capuzzi, B., Palmic, N., Conesa, C., Acker, J., Reguera, J., Lesage, P., Fernandez-Tornero, C.(2023) Nat Commun 14: 1729-1729
- PubMed: 36977686
- DOI: https://doi.org/10.1038/s41467-023-37109-4
- Primary Citation of Related Structures:
7Z0H, 7Z2Z, 7Z30, 7Z31, 8BWS - PubMed Abstract:
The yeast Ty1 retrotransposon integrates upstream of genes transcribed by RNA polymerase III (Pol III). Specificity of integration is mediated by an interaction between the Ty1 integrase (IN1) and Pol III, currently uncharacterized at the atomic level. We report cryo-EM structures of Pol III in complex with IN1, revealing a 16-residue segment at the IN1 C-terminus that contacts Pol III subunits AC40 and AC19, an interaction that we validate by in vivo mutational analysis. Binding to IN1 associates with allosteric changes in Pol III that may affect its transcriptional activity. The C-terminal domain of subunit C11, involved in RNA cleavage, inserts into the Pol III funnel pore, providing evidence for a two-metal mechanism during RNA cleavage. Additionally, ordering next to C11 of an N-terminal portion from subunit C53 may explain the connection between these subunits during termination and reinitiation. Deletion of the C53 N-terminal region leads to reduced chromatin association of Pol III and IN1, and a major fall in Ty1 integration events. Our data support a model in which IN1 binding induces a Pol III configuration that may favor its retention on chromatin, thereby improving the likelihood of Ty1 integration.
Organizational Affiliation:
Centro de Investigaciones Biológicas Margarita Salas, CSIC, 28040, Madrid, Spain.